Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | John Gubatan | -- | 2304 | 2022-07-12 02:18:50 | | | |
| 2 | Camila Xu | Meta information modification | 2304 | 2022-07-12 03:04:10 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Gubatan, J.; Boye, T.L.; Temby, M.; Sojwal, R.S.; Holman, D.R.; Sinha, S.R.; Rogalla, S.R.; Nielsen, O.H. Microbiome in the Pathogenesis of Inflammatory Bowel Disease. Encyclopedia. Available online: https://encyclopedia.pub/entry/25028 (accessed on 07 February 2026).
Gubatan J, Boye TL, Temby M, Sojwal RS, Holman DR, Sinha SR, et al. Microbiome in the Pathogenesis of Inflammatory Bowel Disease. Encyclopedia. Available at: https://encyclopedia.pub/entry/25028. Accessed February 07, 2026.
Gubatan, John, Theresa Louise Boye, Michelle Temby, Raoul S. Sojwal, Derek R. Holman, Sidhartha R. Sinha, Stephan R. Rogalla, Ole Haagen Nielsen. "Microbiome in the Pathogenesis of Inflammatory Bowel Disease" Encyclopedia, https://encyclopedia.pub/entry/25028 (accessed February 07, 2026).
Gubatan, J., Boye, T.L., Temby, M., Sojwal, R.S., Holman, D.R., Sinha, S.R., Rogalla, S.R., & Nielsen, O.H. (2022, July 12). Microbiome in the Pathogenesis of Inflammatory Bowel Disease. In Encyclopedia. https://encyclopedia.pub/entry/25028
Gubatan, John, et al. "Microbiome in the Pathogenesis of Inflammatory Bowel Disease." Encyclopedia. Web. 12 July, 2022.
Copy Citation
The microbiome of patients with inflammatory bowel disease (IBD) is characterized by bacterial dysbiosis (i.e., an imbalance of pathogenic and commensal bacteria). Bacterial diversity has been shown to be reduced during active inflammation in IBD.
gut microbiome
inflammatory bowel disease
ulcerative colitis
1. Microbiome in the Pathogenesis of Inflammatory Bowel Disease (IBD)
The individual human lives in symbiosis with 100 trillion microbiota of the gastrointestinal tract, comprising more than 1000 different types, which are distributed among the genera; bacteria, bacteriophages (bacterial viruses), fungi, and protozoa [1]. The fungal and protozoan microbiome implications in IBD are, however, poorly described [2]. Sparse studies suggest that the protozoan microbiome in patients with active IBD display an increased prevalence of Blastocystis compared to those with quiescent disease or control subjects [3][4]. Additionally, studies report alterations in the diversity (measure of the number of species in a community, and a measure of the abundance of each species) and composition of the fungal microbiome among patients with IBD compared to healthy subjects [5][6][7][8][9][10], and, moreover, intra-individual changes in the fungal composition between inflamed and noninflamed mucosa have been observed in patients with Crohn’s disease (CD) [11]. Intestinal bacteria with associated bacteriophages and the intestinal epithelial cell layer are increasingly being studied, and exist in a dynamic tripartite—both mutualistic and parasitic—relationship, which recently started to be unraveled (Figure 1). Pattern recognition receptors (PRRs) specialized in recognizing bacteria and bacterial products are found in both immune and intestinal epithelial cells. In this way, intestinal epithelial cells balance the composition and luminal microbiota by regulating the secretion of mucus, antimicrobial peptides, and immune mediators, e.g., mucosal immunoglobulin A (IgA) [12]. Nevertheless, surprising evidence also points towards direct communication between bacteriophages and intestinal epithelial cells by bacteriophages adhering to mucosal surfaces, apical-to-basolateral transcytosis (i.e., endocytosis followed by exocytosis transporting bacteriophages across epithelial cells), and by the direct delivery of proteins and nucleic acids to eukaryotic cells [13].
1.1. Bacteria in IBD
The microbiome of patients with IBD is characterized by bacterial dysbiosis (i.e., an imbalance of pathogenic and commensal bacteria). Bacterial diversity has been shown to be reduced during active inflammation in IBD [14][15]. Furthermore, gut microbiome composition has been shown to vary based on their location along the gastrointestinal tract [16]. This observation is probably driven by mucosal changes in tissue oxygenation and disruption of the mucosal barrier function in IBD [15]. Bacterial dysbiosis, which refers to an imbalance of pathogenic and commensal bacteria, is in IBD characterized by a depletion of the phyla Actinobacteria, Firmicutes, and Bacteroidetes [17][18][19][20], and an enrichment of Proteobacteria [21]. Interestingly, Firmicutes and Bacteroidetes are primary producers of energy substrates for intestinal epithelial cells and anti-inflammatory agents, including butyrate and other short-chain fatty acids (SCFAs) [22][23]. Not surprisingly, fecal samples of patients with IBD display a decreased amount of SCFAs [24]. Moreover, long-term remission normalizes both the bacterial microbiota and SCFAs levels in a majority of IBD patients, although with pronounced interindividual variations [25][26][27]. Additionally, low levels of Firmicutes and Faecalibacterium species appear to be related to a high risk of relapse and post-operative recurrence of IBD patients [28][29][30][31]. Polymorphisms of the NOD2 gene are associated with an abundance of Faecalibacterium prausnitzii, the Roseburia genus and the Enterobacteriaceae family [32][33]. Additionally, the microbiome is affected by the diet of the host [34][35]. Interestingly, the intake of prebiotics such as nondigestible fibers is positively correlated with circulating serum levels of granulocyte-macrophage colony stimulating factor (GM-CSF) and negatively correlated with interleukin (IL)-6 and IL-8. These cytokines play central roles in the pathogenesis of IBD [36] and could be a result of altered bacteria or bacterial metabolites in the intestinal lumen. Thus, an intimate relationship between host bacterial microbiome and epithelial cells is evident in the pathogenesis of IBD. Hence, bacteria or bacterial products regulate components of the immune system, but an intestinal chronic low-grade inflammatory environment causing tissue oxygenation and disruption of the mucosal barrier may, on the other hand, significantly impact the microbiome by selecting against inflammatory sensitive species and inducing blooms in evolutionary adapted species.
1.2. Bacteriophages in IBD
The virome of the gut is dominated by viruses that infect bacteria, the so-called bacteriophages (phages), that can present themselves as RNA or both double- and single-stranded DNA [37]. Thus, patients with IBD display an elevated intestinal phage diversity and abundance [38][39]. Importantly, this expansion and diversification of the intestinal bacteriophages is not secondary to the observed concomitant and significantly reduced bacterial diversity [39].
Bacteriophages can indirectly stimulate the immune system by mediating bacterial lysis, which subsequently cause the release of phosphorus-containing bacterial components along with active enzymes [40], but they can also be directly sensed by intestinal epithelial cells and innate immune cells. In fact, bacteriophages have recently been found to be embedded within the intestinal mucus, and are transported across the intestinal epithelial barrier via transcytosis [13].
One of the major obstacles to comprehensively defining the virome is “viral dark matter”, i.e., metagenomic sequences originating from viruses, which, do not align with any reference virus sequences [41]. This is caused by a lack of universal marker genes on phages (similar to the 16S ribosomal RNA gene in bacteria or the 18S and internal transcriptional spacer (ITS) ribosomal RNA genes in eukaryotes), a lack of taxonomic information due to poorly populated databases, and the fact that the virome exhibits an enormous diversity and interindividual variation [42]. Additionally, bacteriophages remain hard to culture and are challenging to analyze. Nevertheless, recent data using whole-virome analysis have shed some light on the viral dark matter in IBD [43]. Intestinal bacteriophages exist in two states: lytic or temperate. The lytic cycle results in destruction of the infected cells, and the temperate phages integrate their genomes into their host bacterial chromosome [44].
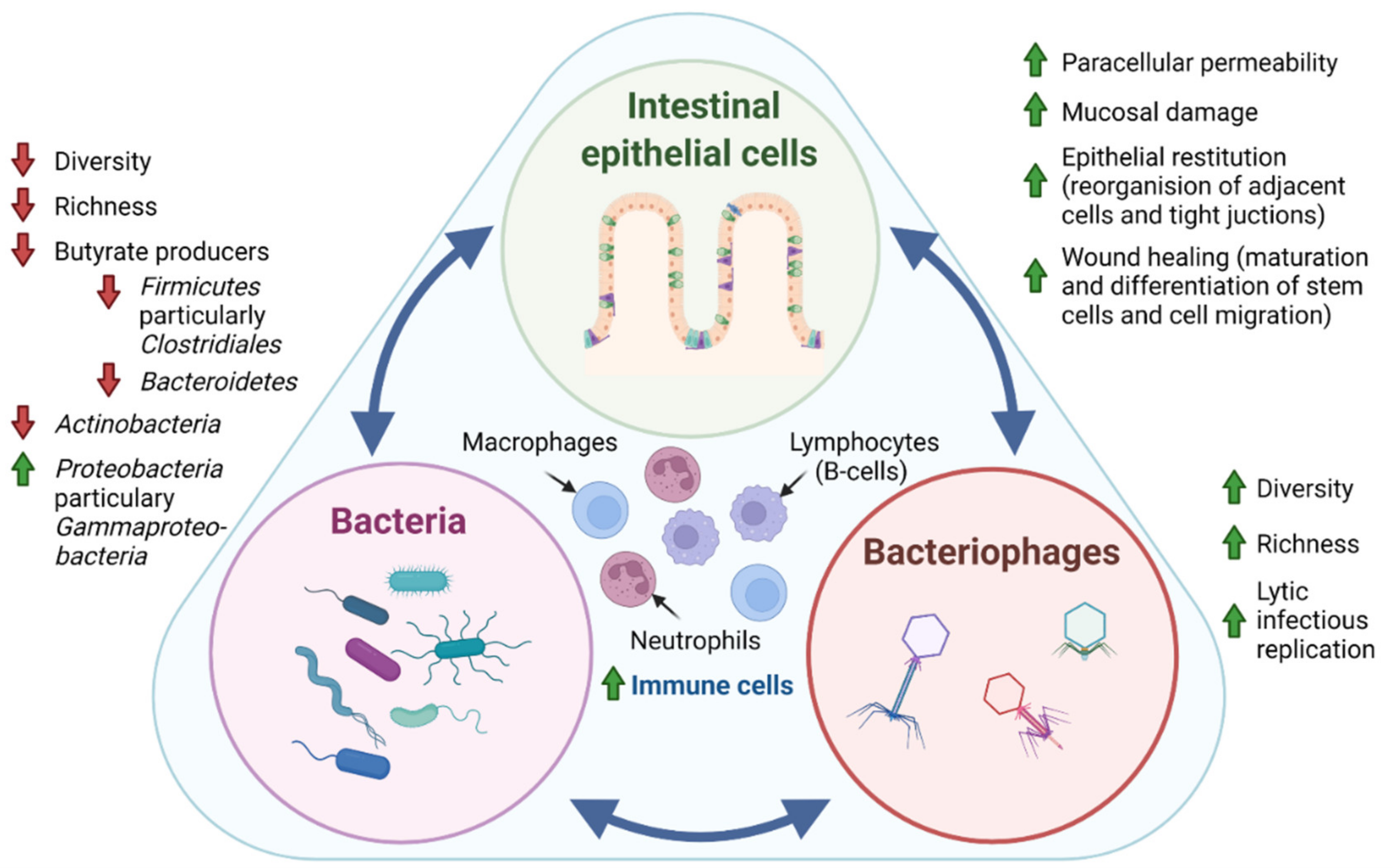 Figure 1. Tripartite relationship between the intestinal epithelial cells, bacteria, and bacteriophages in IBD pathogenesis. In IBD pathogenesis, bacterial dysbiosis is characterized by decreased bacterial diversity (measure of the number of species in a community, and a measure of the abundance of each species) and richness (number of species in a community) evident by the depletion of the phyla Actinobacteria, Firmicutes, and Bacteroidetes and an enrichment of Proteobacteria. In contrast, studies generally suggest that intestinal bacteriophages, which are viruses that infect and replicate within bacteria, display increased diversity and richness. Interestingly, it has recently been suggested that the temperate phage population displays a shift from lysogenic to lytic replication in patients with IBD [43]. Where intestinal epithelial cells are known to directly regulate the secretion of mucus, antimicrobial peptides, and immune mediators through patterns recognition receptors (PRR), surprising evidence also points towards direct communication between bacteriophages and epithelial cells by adhering to mucosal surfaces, apical-to-basolateral transcytosis, and by the direct delivery of proteins and nucleic acids to eukaryotic cells. Thus, the intestinal epithelial cell layer, intestinal bacteria, and bacteriophages exist in a dynamic tripartite—both mutualistic and parasitic—relationship. Further, sparse studies propose that fungal and protozoan microbiomes are also affected in IBD pathogenesis, displaying both altered diversity and composition. The mechanistic interplay between intestinal epithelial cells, bacteria, bacteriophages, as well as fungi and protozoa, has yet to be unraveled, but would potentially provide insight for future clinical applications of microbiota in IBD. Green arrow: increased, red arrow: decreased.
Figure 1. Tripartite relationship between the intestinal epithelial cells, bacteria, and bacteriophages in IBD pathogenesis. In IBD pathogenesis, bacterial dysbiosis is characterized by decreased bacterial diversity (measure of the number of species in a community, and a measure of the abundance of each species) and richness (number of species in a community) evident by the depletion of the phyla Actinobacteria, Firmicutes, and Bacteroidetes and an enrichment of Proteobacteria. In contrast, studies generally suggest that intestinal bacteriophages, which are viruses that infect and replicate within bacteria, display increased diversity and richness. Interestingly, it has recently been suggested that the temperate phage population displays a shift from lysogenic to lytic replication in patients with IBD [43]. Where intestinal epithelial cells are known to directly regulate the secretion of mucus, antimicrobial peptides, and immune mediators through patterns recognition receptors (PRR), surprising evidence also points towards direct communication between bacteriophages and epithelial cells by adhering to mucosal surfaces, apical-to-basolateral transcytosis, and by the direct delivery of proteins and nucleic acids to eukaryotic cells. Thus, the intestinal epithelial cell layer, intestinal bacteria, and bacteriophages exist in a dynamic tripartite—both mutualistic and parasitic—relationship. Further, sparse studies propose that fungal and protozoan microbiomes are also affected in IBD pathogenesis, displaying both altered diversity and composition. The mechanistic interplay between intestinal epithelial cells, bacteria, bacteriophages, as well as fungi and protozoa, has yet to be unraveled, but would potentially provide insight for future clinical applications of microbiota in IBD. Green arrow: increased, red arrow: decreased.1.3. Clinical Relevance of Gut Microbiota in IBD
Previously, it was believed that patients with CD would benefit from antibiotic therapies, resulting in a deleterious effect on the intestinal microbiota [45]. Nonetheless, exposure with antibiotics has been associated with increased microbial dysbiosis [46], and no scientific evidence exists for a beneficial effect of the antibiotic treatment of patients with CD without fistulas or ongoing infections. Instead, an increasing number of clinical trials have been initiated with the aim of investigating the therapeutic effect of fecal microbiota transplantation (FMT) in patients with IBD [47][48].
In the largest, double-blind, randomized, placebo-controlled clinical trial of donor FMT for UC to date, the primary outcome was defined as steroid-free clinical remission with endoscopic remission or response. The primary outcome was achieved in 11 (27%) of 41 patients allocated FMT versus three (8%) of 40 who were assigned placebo (p < 0.04) [49]. Another study of adults with mild to moderate UC compared anaerobically prepared pooled donor FMT versus autologous FMT. Here, 12 of the 38 participants (32%) receiving pooled donor FMT, as compared to 3 of the 35 (9%) receiving autologous FMT, experienced an 8-week steroid-free clinical remission (p < 0.03) [50]. These studies and other published data indicate that donor FMT induces remission in a statistically significant proportion of UC patients [49][50][51][52].
However, available studies in patients with CD are scarce and under-powered. One study with adult colonic or ileo-colonic CD (n = 17, 8 FMT and 9 sham) showed that the steroid-free clinical remission rate at 10 and 24 weeks was 4 of 9 (44%) and 3 of 9 (33%), respectively, in the sham transplantation group and 7 of 8 (88%) and 4 of 8 (50%) in the FMT group (p > 0.05 at both time points), and none of the patients reached the primary endpoint [53]. These results are currently being tested in a larger ongoing clinical trial (n = 24) (ClinicalTrials.gov identifier NCT02097797). Interestingly, several studies in both UC and CD have revealed a significant shift in fecal microbial composition towards a greater microbial diversity, like that of healthy subjects in patients who experienced clinical responses [52][54][55][56]. Although FMT studies to date report low FMT-associated adverse effects in UC [51][55], one study reported flares within a few days of undergoing FMT in CD [56]. Thus, further research is warranted to assess the long-term maintenance of remission and safety of both donor and therapeutically optimized donor-derived strains [57].
2. Dietary Modulation of the Gut Microbiome in IBD
Interests in the diet’s ability to alter the gut microbiome as a therapeutic strategy among patients with IBD has grown tremendously in recent years (Figure 2) [58]. Several nutritional therapies have been explored among pediatric patients with IBD. For example, the Crohn’s Disease Exclusion Diet (CDED) is a high protein, low-fat diet that includes foods such as chicken, fish, eggs, rice, potatoes, and various fruits and vegetables. This intervention has been effective for mild to moderate CD in children, as well as for patients whose response to anti-TNF biologic treatments plateaued [58]. Levine et al. found that remission in the CDED groups was associated with changes in microbial diversity, a decrease in Proteobacteria and an increase in Firmicutes, particularly Clostridiales. Remission also led to a significant decrease in Proteobacteria, particularly Gammaproteobacteria [59].
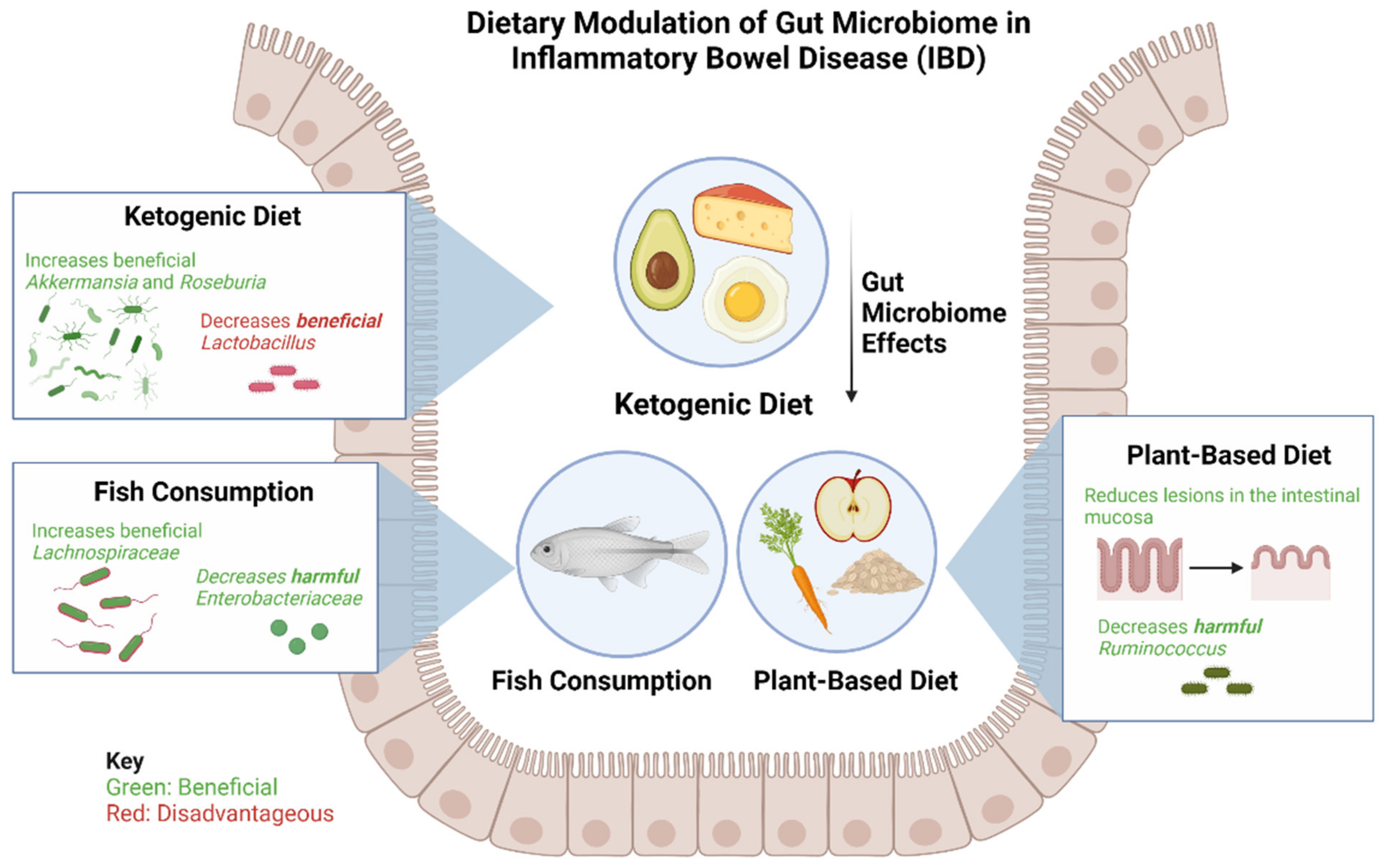 Figure 2. Effects of a ketogenic diet, plant-based diet, and fish consumption on gut microbiome in patients with IBD. The ketogenic diet has been shown to increase beneficial bacteria Akkermansia and Roseburia and consequently decrease beneficial Lactobacillus. The plant-based diet has been found to be beneficial in reducing lesions of the intestinal mucosa and reducing harmful Ruminococcus. Fish consumption leads to an increase in beneficial Lachnospiraceae and a decrease in harmful Enterobacteriaceae.
Figure 2. Effects of a ketogenic diet, plant-based diet, and fish consumption on gut microbiome in patients with IBD. The ketogenic diet has been shown to increase beneficial bacteria Akkermansia and Roseburia and consequently decrease beneficial Lactobacillus. The plant-based diet has been found to be beneficial in reducing lesions of the intestinal mucosa and reducing harmful Ruminococcus. Fish consumption leads to an increase in beneficial Lachnospiraceae and a decrease in harmful Enterobacteriaceae.A Ketogenic Diet (KD) and Low-Carbohydrate Diet (LCD) show promising changes in the specific composition and function of gut microbiota and metabolites in mice [60]. The study by Kong et al. found that, after inducing colitis, KD significantly reduced inflammatory responses, protected intestinal barrier function, and reduced the expression of inflammatory cytokines, whereas the opposite effects were observed for the LCD [60]. These findings indicate a promising dietary strategy for treating IBD, and demonstrate, for the first time, that fecal microbiota transplantation from donors on a KD confers microbiota benefits and relieves colitis in dextran sulfate sodium (DSS)-induced recipients [59]. KD dramatically increased the abundance of Akkermansia and Roseburia; expanding the abundance of Akkermansia has been associated with improved glucose homeostasis, modulated immune responses, and protected barrier function [60]. It should, however, be noted that while KD alleviated the progression of intestinal inflammation, it also reduced the abundance of some healthy bacteria, such as Lactobacillus, compared with a normal diet [60].
Fiber in fruits and vegetables has been shown to provide several benefits to patients with IBD, such as prolonging remission and reducing lesions in the intestinal mucosa, while an imbalance in the consumption of fiber is a risk factor for IBD development [61]. Furthermore, a diet rich in oats prevents the worsening of gastrointestinal symptoms in UC, while a diet rich in high-fiber legumes mitigates intestinal inflammation in rodent models of IBD [61].
Alternatively, fish consumption can lower the risk of IBD. Studies have found ω3FAs to support anti-inflammatory processes when interacting with microbes and alter microbiota diversity, increase beneficial bacteria, and reduce harmful bacteria. ω3FAs encourage growth of SCFA-producing microbes, including the Lachnospiraceae, and lessen the abundance of pathogenic microbes, such as Enterobacteriaceae, in infants [61]. However, the exact pathways and interactions between ω3FAs and the microbes themselves remain unclear.
As interest grows in the benefits of a plant-based diet in IBD, studies have demonstrated that processed and animal-derived foods, in contrast, are associated with higher abundances of CD and UC inflammatory species such as Ruminococcus, as well as with an elevated calprotectin, the gut-specific inflammatory marker [62]. Allin et al. found that processed meat, soft drinks, refined sweetened foods, and salty foods are associated with a higher risk of developing IBD [63]. The study associates the excessive ingestion of ultra-processed foods (UPF) with an increased risk of IBD. Thus, compared with one serving of UPF per day, 5 or more servings per day was associated with a hazard ratio of IBD of 1.82 (95% confidence interval, 1.22–2.72). Unprocessed foods, such as white meat, dairy, starch, fruit, vegetables, and legumes, were shown not to be associated with IBD, while fried foods were associated with a higher rate of both CD and UC. IBD development is not affected by individual food categories (meats, dairy, starch, and fruit and vegetables), suggesting that consuming overly processed foods may be a major factor in diet-related IBD development [64].
Why and how processed/animal-derived foods in the gut may cause inflammation is still unknown. However, it is suggested that the processed sugars, red meats, and saturated fats abundant in the Western diet drastically alter the tissue and barrier function of the intestines, which trigger an inflammatory response leading to an imbalance of the TH17/Treg axis [64]. It is currently unknown whether Western diets also lead to adverse IBD outcomes in patients with well-established CD or UC; thus, a gap exists, which future studies must investigate.
References
- Almeida, A.; Mitchell, A.L.; Boland, M.; Boland, M.; Forster, S.; Gloor, G.; Tarkowska, A.; Lawley, T.D.; Finn, R.D. A new genomic blueprint of the human gut microbiota. Nature 2019, 568, 499–504.
- Guzzo, G.L.; Andrews, J.M.; Weyrich, L.S. The Neglected Gut Microbiome: Fungi, Protozoa, and Bacteriophages in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2022, 28, 1112–1122.
- Petersen, A.M.; Stensvold, C.R.; Mirsepasi, H.; Engberg, J.; Friis-Møller, A.; Porsbo, L.J.; Hammerum, A.M.; Nordgaard-Lassen, I.; Nielsen, H.V.; Krogfelt, K.A. Active ulcerative colitis associated with low prevalence of Blastocystis and Dientamoeba fragilis infection. Scand. J. Gastroenterol. 2013, 48, 638–639.
- Tito, R.Y.; Chaffron, S.; Caenepeel, C.; Lima-Mendez, G.; Wang, J.; Vieira-Silva, S.; Falony, G.; Hildebrand, F.; Darzi, Y.; Rymenans, L.; et al. Population-level analysis of Blastocystis subtype prevalence and variation in the human gut microbiota. Gut 2019, 68, 1180–1189.
- Chehoud, C.; Albenberg, L.G.; Judge, C.; Hoffmann, C.; Grunberg, S.; Bittinger, K.; Baldassano, R.N.; Lewis, J.D.; Bushman, F.D.; Wu, G.D. Fungal Signature in the Gut Microbiota of Pediatric Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2015, 21, 1948–1956.
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048.
- Hoarau, G.; Mukherjee, P.K.; Gower-Rousseau, C.; Hager, C.; Chandra, J.; Retuerto, M.A.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.F.; et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. MBio 2016, 7, e01250-16.
- Qiu, X.; Ma, J.; Jiao, C.; Mao, X.; Zhao, X.; Lu, M.; Wang, K.; Zhang, H. Alterations in the mucosa-associated fungal microbiota in patients with ulcerative colitis. Oncotarget 2017, 8, 107577–107588.
- Liguori, G.; Lamas, B.; Richard, M.L.; Brandi, G.; Da Costa, G.; Hoffmann, T.W.; Di Simone, M.P.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Fungal Dysbiosis in Mucosa-associated Microbiota of Crohn’s Disease Patients. J. Crohns Colitis 2016, 10, 296–305.
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237.
- Jain, U.; Ver Heul, A.M.; Xiong, S.; Gregory, M.H.; Demers, E.G.; Kern, J.T.; Lai, C.-W.; Muegge, B.D.; Barisas, D.A.G.; Leal-Ekman, J.S.; et al. Debaryomyces is enriched in Crohn’s disease intestinal tissue and impairs healing in mice. Science 2021, 371, 1154–1159.
- Abraham, C.; Abreu, M.T.; Turner, J.R. Pattern Recognition Receptor Signaling and Cytokine Networks in Microbial Defenses and Regulation of Intestinal Barriers: Implications for Inflammatory Bowel Disease. Gastroenterology 2022, 162, 1602–1616.e6.
- Nguyen, S.; Baker, K.; Padman, B.S.; Patwa, R.; Dunstan, R.A.; Weston, T.A.; Schlosser, K.; Bailey, B.; Lithgow, T.; Lazarou, M.; et al. Bacteriophage Transcytosis Provides a Mechanism to Cross Epithelial Cell Layers. MBio 2017, 8, e01874-17.
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211.
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392.
- Sepehri, S.; Kotlowski, R.; Bernstein, C.N.; Krause, D.O. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm. Bowel Dis. 2007, 13, 675–683.
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662.
- Michail, S.; Durbin, M.; Turner, D.; Griffiths, A.M.; Mack, D.R.; Hyams, J.; Leleiko, N.; Kenche, H.; Stolfi, A.; Wine, E. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm. Bowel Dis. 2012, 18, 1799–1808.
- Alam, M.T.; Amos, G.C.A.; Murphy, A.R.J.; Murch, S.; Wellington, E.M.H.; ArJasaradnam, R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020, 12, 1.
- Zhou, Y.; Zhi, F. Lower Level of Bacteroides in the Gut Microbiota Is Associated with Inflammatory Bowel Disease: A Meta-Analysis. Biomed Res. Int. 2016, 2016, 5828959.
- Vester-Andersen, M.K.; Mirsepasi-Lauridsen, H.C.; Prosberg, M.V.; Mortensen, C.O.; Träger, C.; Skovsen, K.; Thorkilgaard, T.; Nøjgaard, C.; Vind, I.; Krogfelt, K.A.; et al. Increased abundance of proteobacteria in aggressive Crohn’s disease seven years after diagnosis. Sci. Rep. 2019, 9, 13473.
- Stoeva, M.K.; Garcia-So, J.; Justice, N.; Myers, J.; Tyagi, S.; Nemchek, M.; McMurdie, P.J.; Kolterman, O.; Eid, J. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes 2021, 13, 1907272.
- Chia, L.W.; Mank, M.; Blijenberg, B.; Aalvink, S.; Bongers, R.S.; Stahl, B.; Knol, J.; Belzer, C. Bacteroides thetaiotaomicron Fosters the Growth of Butyrate-Producing Anaerostipes caccae in the Presence of Lactose and Total Human Milk Carbohydrates. Microorganisms 2020, 8, 1513.
- Huda-Faujan, N.; Abdulamir, A.S.; Fatimah, A.B.; Anas, O.M.; Shuhaimi, M.; Yazid, A.; Loong, Y. The impact of the level of the intestinal short chain Fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem. J. 2010, 4, 53–58.
- Stange, E.F.; Schroeder, B.O. Microbiota and mucosal defense in IBD: An update. Expert Rev. Gastroenterol. Hepatol. 2019, 13, 963–976.
- Kumari, R.; Ahuja, V.; Paul, J. Fluctuations in butyrate-producing bacteria in ulcerative colitis patients of North India. World J. Gastroenterol. 2013, 19, 3404–3414.
- Wills, E.S.; Jonkers, D.M.; Savelkoul, P.H.; Masclee, A.A.; Pierik, M.J.; Penders, J. Fecal microbial composition of ulcerative colitis and Crohn’s disease patients in remission and subsequent exacerbation. PLoS ONE 2014, 9, e90981.
- Rajca, S.; Grondin, V.; Louis, E.; Vernier-Massouille, G.; Grimaud, J.C.; Bouhnik, Y.; Laharie, D.; Dupas, J.-L.; Pillent, H.; Picon, L.; et al. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn’s disease. Inflamm. Bowel Dis. 2014, 20, 978–986.
- Braun, T.; Di Segni, A.; BenShoshan, M.; Neuman, S.; Levhar, N.; Bubis, M.; Picard, O.; Sosnovski, K.; Efroni, G.; Farage Barhom, S.; et al. Individualized Dynamics in the Gut Microbiota Precede Crohn’s Disease Flares. Am. J. Gastroenterol. 2019, 114, 1142–1151.
- Wright, E.K.; Kamm, M.A.; Wagner, J.; Teo, S.-M.; De Cruz, P.; Hamilton, A.L.; Ritchie, K.J.; Inouye, M.; Kirkwood, C.D. Microbial Factors Associated with Postoperative Crohn’s Disease Recurrence. J. Crohns Colitis 2017, 11, 191–203.
- Varela, E.; Manichanh, C.; Gallart, M.; Torrejón, A.; Borruel, N.; Casellas, F.; Guarner, F.; Antolin, M. Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Aliment. Pharmacol. Ther. 2013, 38, 151–161.
- Aschard, H.; Laville, V.; Tchetgen, E.T.; Knights, D.; Imhann, F.; Seksik, P.; Zaitlen, N.; Silverberg, M.S.; Cosnes, J.; Weersma, R.K.; et al. Genetic effects on the commensal microbiota in inflammatory bowel disease patients. PLoS Genet. 2019, 15, e1008018.
- Knights, D.; Silverberg, M.S.; Weersma, R.K.; Gevers, D.; Dijkstra, G.; Huang, H.; Tyler, A.D.; Van Sommeren, S.; Imhann, F.; Stempak, J.M.; et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014, 6, 107.
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS ONE 2013, 8, e66019.
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563.
- Olendzki, B.; Bucci, V.; Cawley, C.; Maserati, R.; McManus, M.; Olednzki, E.; Madziar, C.; Chiang, D.; Ward, D.V.; Pellish, R.; et al. Dietary manipulation of the gut microbiome in inflammatory bowel disease patients: Pilot study. Gut Microbes 2022, 14, 2046244.
- Duan, Y.; Young, R.; Schnabl, B. Bacteriophages and their potential for treatment of gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 135–144.
- Wang, W.; Jovel, J.; Halloran, B.; Wine, E.; Patterson, J.; Ford, G.; O’keefe, S.; Meng, B.; Song, D.; Zhang, Y.; et al. Metagenomic analysis of microbiome in colon tissue from subjects with inflammatory bowel diseases reveals interplay of viruses and bacteria. Inflamm. Bowel Dis. 2015, 21, 1419–1427.
- Norman, J.M.; Handley, S.A.; Baldridge, M.T.; Droit, L.; Liu, C.Y.; Keller, B.C.; Kambal, A.; Monaco, C.L.; Zhao, G.; Fleshner, P.; et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015, 160, 447–460.
- Heyer, C.M.; Weiss, E.; Schmucker, S.; Rodehutscord, M.; Hoelzle, L.E.; Mosenthin, R.; Stefalnski, V. The impact of phosphorus on the immune system and the intestinal microbiota with special focus on the pig. Nutr. Res. Rev. 2015, 28, 67–82.
- Tisza, M.J.; Pastrana, D.V.; Welch, N.L.; Stewart, B.; Peretti, A.; Starrett, G.J.; Pang, Y.-Y.S.; Krishnamurthy, S.R.; Pesavento, P.A.; McDermott, D.H.; et al. Discovery of several thousand highly diverse circular DNA viruses. eLife 2020, 9, e51971.
- Reyes, A.; Haynes, M.; Hanson, N.; Angly, F.E.; Heath, A.C.; Rohwer, F.; Gordon, J.I. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 2010, 466, 334–338.
- Clooney, A.G.; Sutton, T.D.S.; Shkoporov, A.N.; Holohan, R.K.; Daly, K.M.; O’Regan, O.; Ryan, F.J.; Draper, L.A.; Plevy, S.E.; Ross, R.P.; et al. Whole-Virome Analysis Sheds Light on Viral Dark Matter in Inflammatory Bowel Disease. Cell Host Microbe 2019, 26, 764–778.e765.
- Chen, Y.; Yang, L.; Yang, D.; Song, J.; Wang, C.; Sun, E.; Gu, C.; Chen, H.; Tong, Y.; Tao, P.; et al. Specific Integration of Temperate Phage Decreases the Pathogenicity of Host Bacteria. Front. Cell. Infect. Microbiol. 2020, 10, 14.
- Colombel, J.F.; Cortot, A.; van Kruiningen, H.J. Antibiotics in Crohn’s disease. Gut 2001, 48, 647.
- Lewis, J.D.; Chen, E.Z.; Baldassano, R.N.; Otley, A.R.; Griffiths, A.M.; Lee, D.; Bittinger, K.; Bailey, A.; Friedman, E.S.; Hoffmann, C.; et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe 2015, 18, 489–500.
- Allegretti, J.R.; Mullish, B.H.; Kelly, C.; Fischer, M. The evolution of the use of faecal microbiota transplantation and emerging therapeutic indications. Lancet 2019, 394, 420–431.
- Luo, H.; Cao, G.; Luo, C.; Tan, D.; Vong, C.T.; Xu, Y.; Wang, S.; Lu, H.; Wang, Y.; Jing, W. Emerging pharmacotherapy for inflammatory bowel diseases. Pharmacol. Res. 2022, 178, 106146.
- Paramsothy, S.; Kamm, M.A.; Kaakoush, N.O.; Walsh, A.J.; van den Bogaerde, J.; Samuel, D.; Leong, R.W.L.; Connor, S.; Ng, W.; Paramsothy, R.; et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: A randomised placebo-controlled trial. Lancet 2017, 389, 1218–1228.
- Costello, S.P.; Hughes, P.A.; Waters, O.; Bryant, R.V.; Vincent, A.D.; Blatchford, P.; Katsikeros, R.; Makanyanga, J.; Campaniello, M.A.; Mavrangelos, C.; et al. Effect of Fecal Microbiota Transplantation on 8-Week Remission in Patients with Ulcerative Colitis: A Randomized Clinical Trial. JAMA 2019, 321, 156–164.
- Moayyedi, P.; Surette, M.G.; Kim, P.T.; Libertucci, J.; Wolfe, M.; Onischi, C.; Armstrong, D.; Marshall, J.K.; Kassam, Z.; Reinisch, W.; et al. Fecal Microbiota Transplantation Induces Remission in Patients with Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology 2015, 149, 102–109.e106.
- Brezina, J.; Bajer, L.; Wohl, P.; Ďuricová, D.; Hrabák, P.; Novotný, A.; Koželuhová, J.; Lukáš, M.; Mrázek, J.; Fliegerová, K.; et al. Fecal Microbial Transplantation versus Mesalamine Enema for Treatment of Active Left-Sided Ulcerative Colitis-Results of a Randomized Controlled Trial. J. Clin. Med. 2021, 10, 2753.
- Sokol, H.; Landman, C.; Seksik, P.; Berard, L.; Montil, M.; Nion-Larmurier, I.; Bourrier, A.; Le Gall, G.; Lalande, V.; De Rougemont, A.; et al. Fecal microbiota transplantation to maintain remission in Crohn’s disease: A pilot randomized controlled study. Microbiome 2020, 8, 12.
- Vaughn, B.P.; Vatanen, T.; Allegretti, J.R.; Bai, A.; Xavier, R.J.; Korzenik, J.; Gevers, D.; Ting, A.; Robson, S.C.; Moss, A.C. Increased Intestinal Microbial Diversity Following Fecal Microbiota Transplant for Active Crohn’s Disease. Inflamm. Bowel Dis. 2016, 22, 2182–2190.
- Rossen, N.G.; Fuentes, S.; van der Spek, M.J.; Tijssen, J.G.; Hartman, J.H.A.; Duflou, A.; Löwenberg, M.; Van Den Brink, G.R.; Mathus-Vliegen, E.M.H.; de Vos, W.M.; et al. Findings from a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology 2015, 149, 110–118.e114.
- Gutin, L.; Piceno, Y.; Fadrosh, D.; Lynch, K.; Zydek, M.; Kassam, Z.; LaMere, B.; Terdiman, J.; Ma, A.; Somsouk, M.; et al. Fecal microbiota transplant for Crohn disease: A study evaluating safety, efficacy, and microbiome profile. United Eur. Gastroenterol. J. 2019, 7, 807–814.
- Lima, S.F.; Gogokhia, L.; Viladomiu, M.; Chou, L.; Putzel, G.; Jin, W.-B.; Pires, S.; Guo, C.-J.; Gerardin, Y.; Crawford, C.V.; et al. Transferable Immunoglobulin A-Coated Odoribacter splanchnicus in Responders to Fecal Microbiota Transplantation for Ulcerative Colitis Limits Colonic Inflammation. Gastroenterology 2022, 162, 166–178.
- Hart, L.; Verburgt, C.M.; Wine, E.; Zachos, M.; Poppen, A.; Chavannes, M.; Van Limbergen, J.; Pai, N. Nutritional Therapies and Their Influence on the Intestinal Microbiome in Pediatric Inflammatory Bowel Disease. Nutrients 2022, 14, 4.
- Levine, A.; Wine, E.; Assa, A.; Sigall Boneh, R.; Shaoul, R.; Kori, M.; Cohen, S.; Peleg, S.; Shamaly, H.; On, A.; et al. Crohn’s Disease Exclusion Diet Plus Partial Enteral Nutrition Induces Sustained Remission in a Randomized Controlled Trial. Gastroenterology 2019, 157, 440–450.e8.
- Kong, C.; Yan, X.; Liu, Y.; Huang, L.; Zhu, Y.; He, J.; Gao, R.; Kalady, M.F.; Goel, A.; Qin, H.; et al. Ketogenic diet alleviates colitis by reduction of colonic group 3 innate lymphoid cells through altering gut microbiome. Signal Transduct. Target. Ther. 2021, 6, 154.
- O’Mahony, C.; Amamou, A.; Ghosh, S. Diet–Microbiota Interplay: An Emerging Player in Macrophage Plasticity and Intestinal Health. Int. J. Mol. Sci. 2022, 23, 3901.
- Bolte, L.A.; Vich Vila, A.; Imhann, F.; Collij, V.; Gacesa, R.; Peters, V.; Wijmenga, C.; Kurilshikov, A.; Campmans-Kuijpers, M.J.E.; Fu, J.; et al. Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome. Gut 2021, 70, 1287–1298.
- Allin, K.H.; Ungaro, R.C.; Agrawal, M. Ultraprocessed Foods and the Risk of Inflammatory Bowel Disease: Is it Time to Modify Diet? Gastroenterology 2022, 162, 652–654.
- Tracy, M.; Khalili, H. You Are What You Eat? Growing Evidence That Diet Influences the Risk of Inflammatory Bowel Disease. J. Crohns Colitis 2022, 20, jjac025.
More
Information
Subjects:
Gastroenterology & Hepatology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
12 Jul 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




