Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Eleonora Grigoryan | -- | 5530 | 2022-06-27 17:43:31 | | | |
| 2 | Catherine Yang | -1 word(s) | 5529 | 2022-06-30 04:47:00 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Grigoryan, E.N. Embryonic Retina Self-Organization In Vivo. Encyclopedia. Available online: https://encyclopedia.pub/entry/24634 (accessed on 06 March 2026).
Grigoryan EN. Embryonic Retina Self-Organization In Vivo. Encyclopedia. Available at: https://encyclopedia.pub/entry/24634. Accessed March 06, 2026.
Grigoryan, Eleonora N.. "Embryonic Retina Self-Organization In Vivo" Encyclopedia, https://encyclopedia.pub/entry/24634 (accessed March 06, 2026).
Grigoryan, E.N. (2022, June 29). Embryonic Retina Self-Organization In Vivo. In Encyclopedia. https://encyclopedia.pub/entry/24634
Grigoryan, Eleonora N.. "Embryonic Retina Self-Organization In Vivo." Encyclopedia. Web. 29 June, 2022.
Copy Citation
Self-organization is a process that ensures histogenesis of the eye retina. The phenomenon of self-organisation is discussed in the spatiotemporal context and illustrated by key findings during vertebrate retina development in vivo and retinal regeneration in amphibians in situ. Described also are histotypic 3D structures obtained from the disaggregated retinal progenitor cells of birds and retinal 3D organoids derived from the mouse and human pluripotent stem cells.
retina
development
self-organisation
cellular and molecular mechanisms
1. General Concepts
The ontogeny of the vertebrate retina has been a topic of interest to developmental biologists for many decades. The retina’s development in vertebrates begins early, from the time of the division of the anterior neural plate into domains, with the specification of the so-called eye field occurring in the middle of them. The bilateral optic vesicles, being the eye anlagen, are subsequently formed in this region. The retina appears in the posterior wall of the optic cup, which is formed from the optic vesicle through its invagination. Simultaneously, the eye primordium elongates in the posterior part, and its connection to the brain grows narrower, generating the optic stalk (Figure 1) [1][2]. A lineage tracing and live imaging, carried out on zebrafish embryos, allowed a detailed analysis of cell movements, including extended evagination and rim movement [3][4][5]. Retinal progenitor cells (RPCs) in the optic cup undergo active proliferation, producing the prospective retinal pigmented epithelium (RPE) and neural retina (NR) [1][2]. Those cells that remain in the outer layer of the optic cup constitute the RPE progenitors, whereas the inner layer is composed of the NR progenitors. Each of the RPCs is multipotent and capable of producing the full range of retinal cell types subsequently. Different clones of RPCs have different combinations of precursors of the major cell types, including Müller glial cells. It is postulated that the interactions that specify the differentiation pathway of retinal cells occur relatively late in development [6][7][8][9][10].
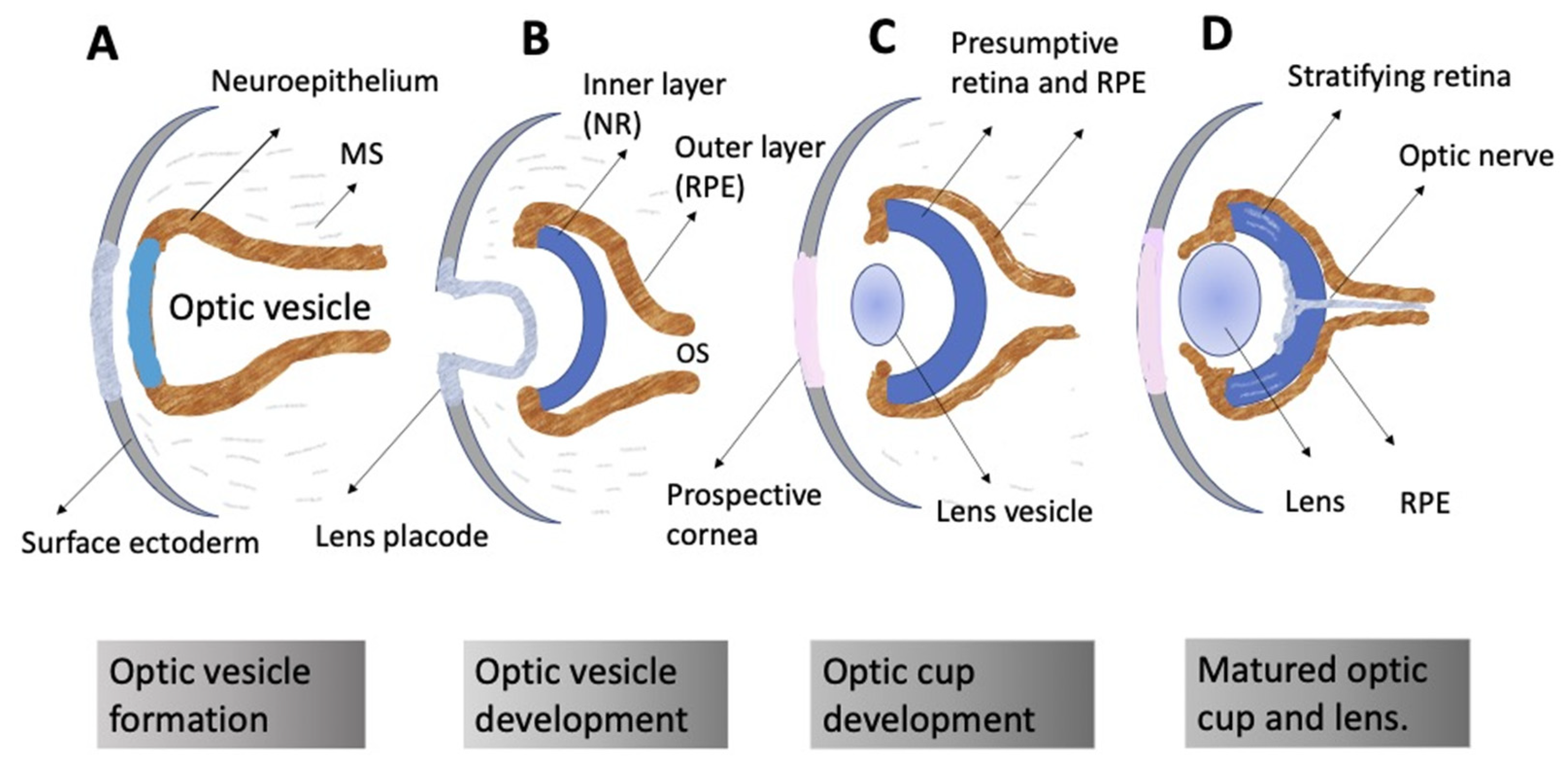
Figure 1. Schematic representation of vertebrate eye development. (A) Formation of the optic vesicle. (B) Specification of the NR (inner layer), RPE (outer layer), and optic stalk (OS) domains within the optic vesicle. Formation of the lens placode from the surface ectoderm. (C) Formation of the optic cup and the lens vesicle. (D) Organization of the formed eye. Neural retina stratification; growth of the optic nerve. MS: mesenchyme.
The “competence model”, based on the sequential maturation of retina cell types in a certain order and as a result of changes in the RPCs’ competencies (potencies), is considered as a fundamental basis for the self-development of the vertebrate NR [9][10][11]. The sequential manner of retinal cell differentiation and its “timing”, exhibiting certain patterns, have been identified in different vertebrate species. It is usually manifested as the generation of ganglion cells, cone photoreceptors, and horizontal and amacrine cells of the retina in the early phase, overlapping with the late-phase generation of rod photoreceptors, bipolar cells, and Müller glia [12]. To date, the main retinal time course, similar to that found in other vertebrates, has been described from the developing human retina using RNA-Seq analysis [13]. The competence model, reflecting the intrinsic pattern of cell diversification in the developing retina, has been further supported by the recently collected extensive information on the expression of transcription factors (TFs) and signaling molecules that jointly determine the hierarchy of RPCs [11][14][15][16][17][18][19] (see below) (Figure 2).
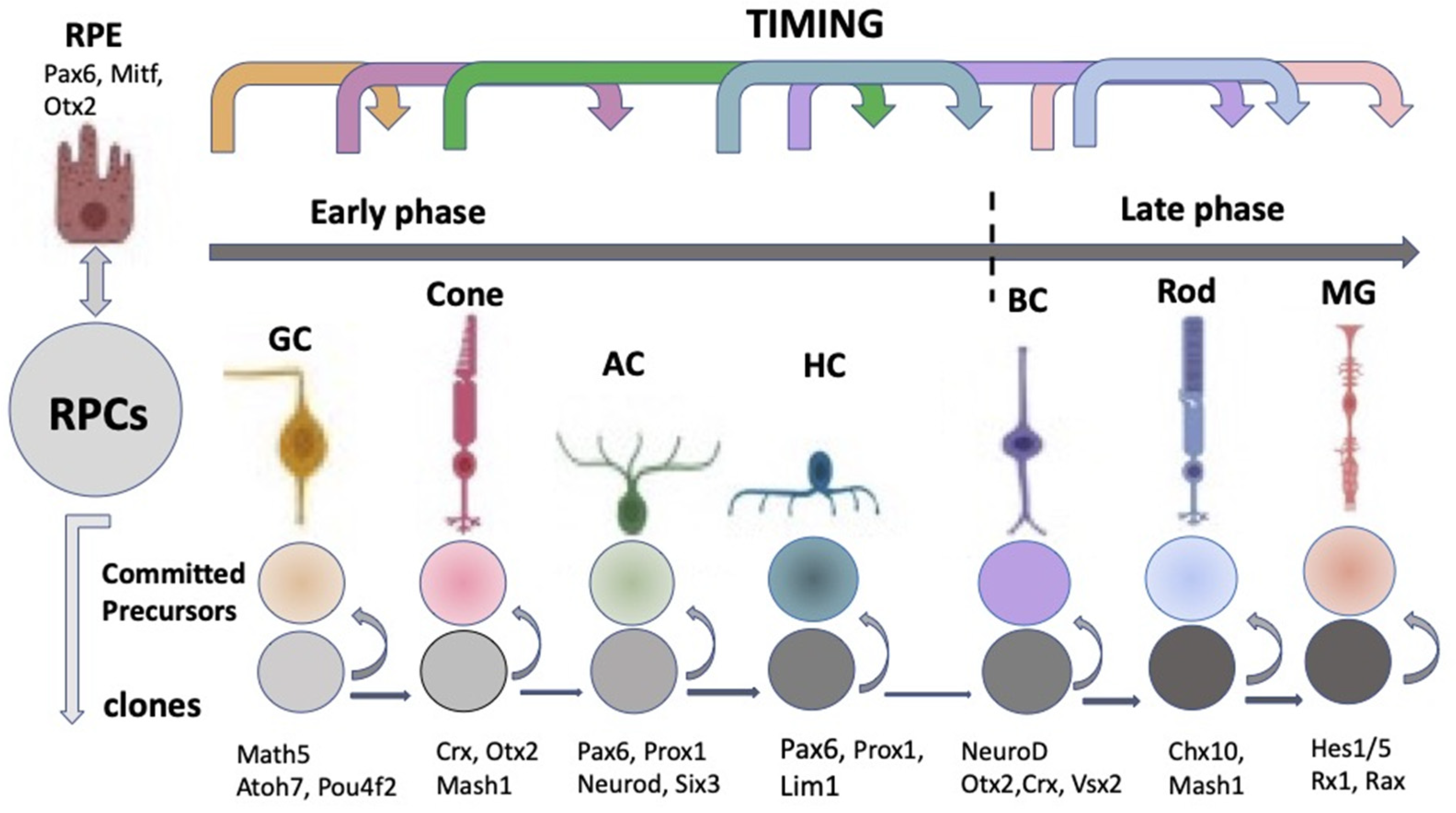
Figure 2. Schematic representation of the classical competence model for eye development. According to the model, retinal progenitor cells progress through the competence windows during which a specific retinal cell type is generated. RPCs—retinal progenitor cells; RPE—retinal pigment epithelium; GC—ganglion cell; AC—amacrine cells; HC—horizontal cells; BC—bipolar cells; MG—Müller glial cell. At the top: the sequential manner of a retinal cell’s differentiation. It has early and late phases. The represented sequence is general for vertebrates, although conditional, since the stages overlap with each other and have species-specific features. Key transcription factors responsible for retinal cell type specification are indicated at the bottom.
The laminar structure of the retina is formed during cell maturation. Its formation in vivo occurs not only through the implementation of internal cell competencies and RPCs’ interactions with each other, but also through external signaling. At the earliest stages, the retina is formed largely through self-organization, a process whose mechanisms are not fully understood. There is some disagreement between the two theories existing today [20]. The first, the above-mentioned competence model, is the theory of RPCs changing competencies, which provides different cell clones for the development of certain cell types at a certain time [12] (Figure 2). This model suggests that during retinogenesis, RPCs acquire and then lose the ability to produce certain cell types in accordance with some internal timing. An assumption has been made that such a behavior of RPCs is regulated and controlled by the cells’ mutual positive/negative feedback through intercellular adhesion, communication, and physical factors, as well as signaling from other developing eye tissues [9][10].
However, in vitro studies on developing zebrafish and rat retinal tissue have shown that the “histogenesis of fates” observed in the overall population is accounted for by the variability of cell determination within clones and that cell fate variability among clones is likely to have a partially stochastic explanation [20][21][22]. To confirm this, information is provided that after the elimination of progenitor cells of one or the other cell type in the zebrafish and mouse embryonic retina in vivo, the remaining cells are able to laminate in the correct order [23][24][25]. Thus, lamination of the retina was restored in Chx10, Kip1 double-null mice in the absence of bipolars [23]. In the transgenic zebrafish, an IPL-like neuropil still formed in cellularly simplified retinas consisting of only bipolars and photoreceptors. Remarkably, in this presynaptic-only neuropil, axons of bipolars could still make presynaptic structures and display sublaminar organization of their axonal terminals [24].
There are also data showing that retinal explants, taken at different time points of development and, therefore, containing different populations of RPCs, reproduce in vitro the same size and composition of clones as in vivo [21]. These facts are evidence that both the competence of RPCs and their fate are stochastic to a certain extent and, at the same time, are regulated by some intrinsic mechanisms. This makes it challenging to link the competence state of RPCs to a specific developmental time, as well as to the fully directive action of external regulation. One of the underlying mechanisms of such stochasticity may be the extreme heterogeneity exhibited by RPCs in their expression of TFs [26]. Nevertheless, the selection of the fate of retinal cells is not completely stochastic, since the frequency of some, certain clone compositions, turns out to be higher than one could expect in the case of their completely stochastic development. Gomez and co-authors [21] support this point of view with a mathematical model, which shows that the probability of occurrence of early cell types, such as, in particular, ganglion cell precursors, decreases over time against the background of an increase in the probability of the production of late cell types such as rods, bipolar cells, etc., rather than disappearing completely.
The process of retinal self-organization includes not only the intriguing abilities of RPCs to behave within the framework of the competence model and, at the same time, stochastically, but also a number of other intriguing properties. One of them is the “overproduction” of cells of early cell populations. It has long been known that clones at early time points are usually much larger than later ones [6]. In later developmental phases, overproduction is purposefully eliminated through cell death so that the populations of retinal cell types reach a number adequate for their specific relationships and functions. Programmed cell death, or apoptosis, is known as a common phenomenon in embryonic development and cell differentiation during histogeneses, in particular in the CNS [27][28][29][30][31][32][33][34][39]. According to Vecino and Acera [28], in the case of the retina, the main functions of cell death are as follows: eliminating the neurons that have not established synapses with partner cells (targets); involvement in the retinal laminar structure formation; and eliminating the cells of transit populations. Cell death in the developing NR is described in a number of early studies, with their data summarized in a review by Valenciano et al. [29]. Two waves of cell death are reported for mammals: the first affects the RPC population in the early retinogenesis phase, while the second in the late phase, during synaptogenesis, when the plexiform layers of the NR are formed [28][29][30]. In addition to the above-mentioned phases, Valenciano et al. [29] indicate morphogenic cell death, a programmed cell death related to optic vesicle evagination, optic cup formation, and closure of the optic fissure. Many of the cellular and molecular mechanisms involved in cell death have been elucidated. In particular, many of the molecular triggers underlying programmed cell death have been discovered [31]. The survival/death of both RPCs and retinal neurons in the late stages of retinal development is regulated by families of molecules that carry out both internal and external control of these processes (summarized in [29][31]). Among external signals, Braunger et al. [31] distinguish NF and TGFβ; in the late phase, neurotrophins (BDNF, CNTF, and NF) and immune modulators are considered [29][32][33]. There is evidence of the role of cell death in the formation of cell mosaicism [34]. The resulting retinal mosaics constitute the “functional units” necessary for providing the normal functions of the retina [35][36][37].
The cell migration that occurs in the retina along with the processes of cell death and stratification and mosaicism formation aims to achieve the definitive localizations of cells in the NR structure [37][38]. The cell types that emerge after exiting the proliferative phase migrate along the radial (apico-basal) axes of the NR anlage. Tangential migration (perpendicular to the radial axis of the NR) is also known. It is reported to be characteristic of cells emerging in the early phase of retinogenesis. This allows them to move a short distance within their laminar position [37][38]. The processes of neuronal migration, lamination, and mosaicism formation are discussed in detail in a review by Amini et al. [34].
Spatiotemporal, precise orchestrated processes such as the fate choice (competence, differentiation) by cells, excess proliferation, migration, and death, as well as the definitive maturation of cells upon reaching the correct location (in terms of time and position) in the overall NR composition provide adult functionality of the retina. These processes are initially based on a high measure of plasticity of neuroepithelial cells to accommodate the spatiotemporal process while maintaining their tissue integrity and architecture. The self-regulation of plasticity properties against the background of intercellular and physical factors of influence in vivo is described in terms of the expression of genes, TFs, controlled by intrinsic and extrinsic factors of the cells.
2. Regulation of Eye and Retina Development by Extrinsic Factors in Vertebrates In Vivo
The role of intrinsic regulatory factors in response to external signaling systems can be observed at all stages of retinal development in vivo. A number of signaling regulatory molecules emitted from neighboring tissues are identified at the very early stage of eye development. A previous work implicates the extracellular matrix (ECM) as a major player in eye structures’ morphogenesis, with the mechanisms of this influence, however, being poorly understood and the roles of individual ECM proteins not fully defined [39][40]. The eye anlage is surrounded by periocular mesenchyme. However, the study of the effect of its ECM proteins is reported [41] to be complicated by the presence of two sources in the periocular mesenchyme: the neural crest and cells of mesodermal origin, whose specific role is difficult to identify. To understand the role of the neural crest, Bryan et al. [41] analyzed the proteins of its basal membrane. For this, embryos of mutant zebrafish lines were used, in which the basal membrane of the neural crest adjacent to the prospective RPE was destroyed genetically. Most neural crest cells were absent, resulting in optic vesicle cells that moved faster and farther than those in wild-type embryos. Rim movement was impaired in the absence of a complete, continuous basal membrane around the RPE, which resulted in optic cup malformations. A search for a key molecular effector involved in the interaction of the neural crest ECM with the developing eye indicated conserved sulfated monomeric glycoproteins, referred to as nidogens, acting as ECM modulators. The authors [41] suggested that the ECM of the basal membrane is a dynamic substrate capable of regulating cell movements in the early eye anlage, as well as spreading of its outer layer, the RPE. There is an opinion that the periocular mesenchyme, regardless of its source, controls the signaling pathways Hh, TGFβ, and Wnt regulating the eye development in vivo [40][42].
The role of the Wnt and BMP pathways in vertebrate eye development was assessed using inhibitors of these signalings, DKK1 and Noggin (for Wnt and BMP, respectively), and exposure to exogenous IGF1 (Glinka et al., 1997; Lamba et al., 2006) [43][44]. Other results [45] provide evidence that BMP signals in early development inhibit the acquisition of the eye field traits. However, at the stage of the formation of optic vesicles, BMP signals running from the forming lens, on the contrary, are necessary for the maintenance of the eye field character, inhibition of dorsal telencephalic cell identity, and specification of NR cells. The inhibition of WNT and BMP in the developing RPCs derived from ESCs in vitro [44] and the injection of IGF1 mRNA to clawed frog (Xenopus) embryos in vivo [46] induced retinal development. In the latter case, this occurred presumably due to the suppression of the Wnt signaling pathway by IGF [47].
The non-canonical, β-catenin-independent Wnt signaling pathway turned out to be important to form and maintain the eye field and to regulate cellular movements during morphogenesis. The development of the eye field has been shown to be at least partially controlled by Wnt11 and Fz5 through local antagonistic interactions with Wnt/β-catenin signaling, which suppresses retinal identity [48]. In turn, Wnt/β-catenin signaling plays an essential role in multiple developmental processes and has a profound effect on cell proliferation and cell fate determination. Faulty regulation of Wnt/β-catenin signaling results in multiple ocular malformations due to defects in the process of cell fate determination and differentiation [49].
In addition to Wnt, BMP, and IGF1, other factors should also be included in the system of regulation of the early retinal formation stages. It was found that FGF, TGFβ, Notch, Vax, retinoids, and Gas1 are responsible for the diversification and stabilization of the two major visual domains (RPE and NR) in the optic vesicles and eye cup [50][51]. The role of FGF and Wnt signaling has been studied in mice both during RPE and NR formation and in maintaining the properties of the retinal growth zone (ciliary margin (CM)). Using a single-cell analysis, Balasubramanian et al. [52] found that FGF along with Wnt signaling regulate the stem properties of CM cells and their entry into differentiation. FGF promotes Wnt signaling in the CM by stabilizing β-catenin, while Wnt signaling converts the NR into either the CM or the RPE depending on FGF signaling; FGF transforms the RPE to the NR or CM depending on Wnt activity. These data collectively showed that the vertebrate eye develops through a phase transition determined by a combinatorial code of FGF and Wnt signaling [52].
3. Regulation of Eye and Retina Development by Intrinsic Factors in Vertebrates In Vivo
The external signaling influences the expression of TFs, whose differential functioning ensures both early and advanced retinal development [1][2][53][54]. TFs are suggested to be the primary determinants of retinal fate choices (Figure 2 and Figure 3). The expression of genes responsible for the emergence of differences in the competence or, in other words, the potential of RPCs to produce certain cell types determines the processes of cell sequential diversification. It should be noted that TFs, whose differential expression is characteristic of the development of the eye and, in particular, the retina, are conserved across species, and this is certainly a convenience for assessing their role in the eye development mechanism [1][2][54][55].
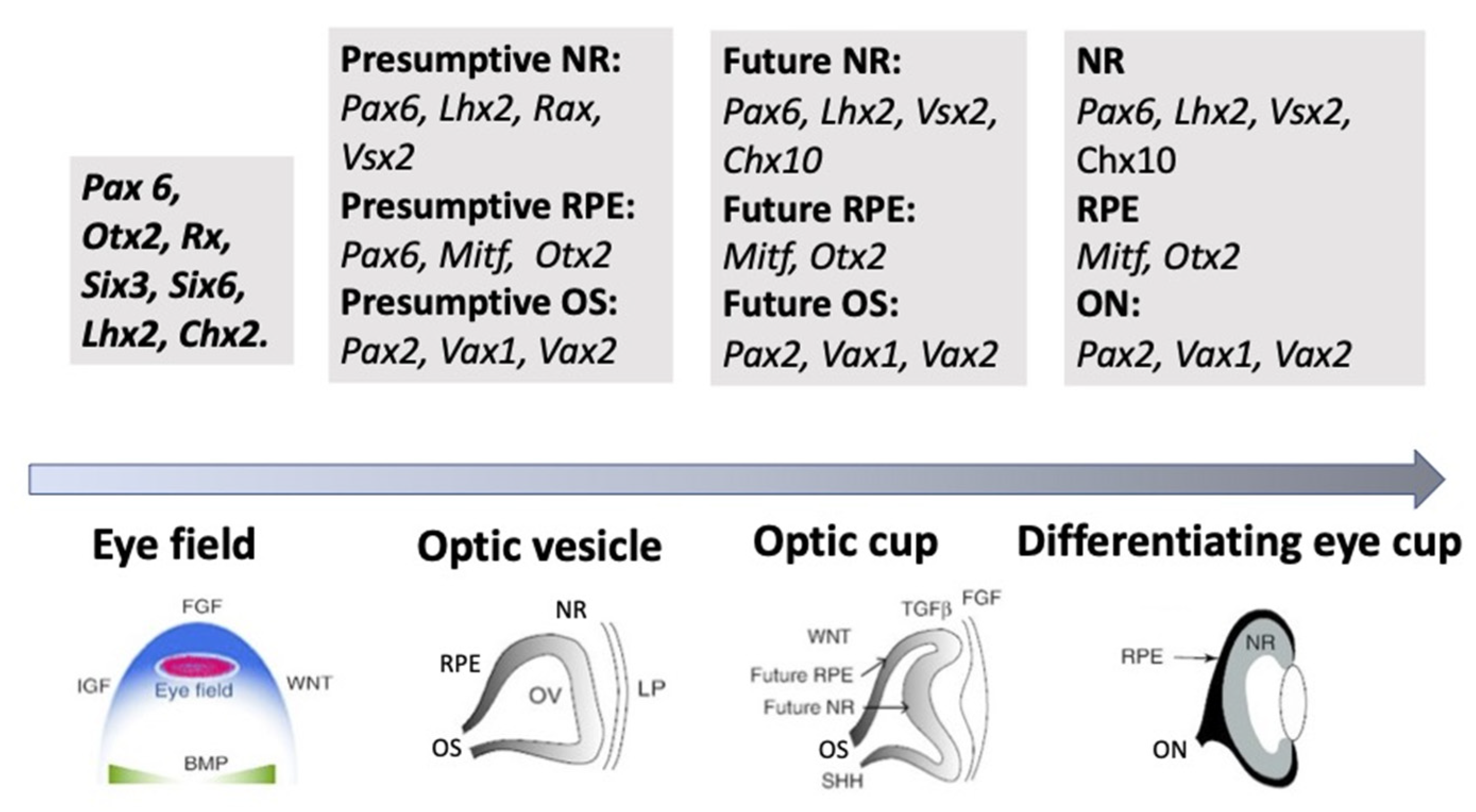
Figure 3. A schematic illustration of the groups of key transcription factors whose expression is associated with the main successive stages of eye development. NR—neural retina, RPE—retinal pigmented epithelium, OS—optic stalk, ON—optic nerve. See the detailed description is in the text.
As mentioned above, the eye’s development is initiated with the emergence of eye field cells in the anterior neural plate during late gastrulation. The formation of this region is accompanied and coordinated by the expression of the eye field TFs (EFTFs). The progenitor cells of the optic vesicle in this region express the same basic genes: Pax6, Rx, Six3,6, Otx2, Sox2, and Lhx2, encoding the respective TFs. In the anterior neural plate, EFTFs compose a genetic network to control the eye specification against the background of inhibition of BMP, Nodal, and Wnt/β-catenin signaling [54][55][56][65,66,67]. The work of TFs in their interaction with signaling pathways at different stages of the eye and retina development (RPE and NR) has been well characterized and presented in many works [1][18][19][53][11,29,30,64]. In brief, it can be represented as follows. As mentioned above, the presence of the eye field in the anterior part of the neural plate is characterized by the expression of EFTFs, in particular Rx. The latter is up-regulated due to the cooperative work of TFs Otx2 and Sox2. The interaction with the BMP, Wnt, and Shh signaling pathways leads to the separation of the eye field area from other areas of the developing brain. Then, the expression of Lhx2 occurs, which allows the initiation of the optic vesicle formation. The expression of TFs Otx2 and Mitf, induced by the TGFβ pathway, as well as Pax6 is characteristic of the optic vesicle in general. However, when the prospective NR is formed, Mitf is down-regulated with the involvement of Vsx2. The formation of RPE requires the suppression of FGF signaling, as well as Sox2, which occurs with the involvement of Otx2. Thus, the signaling involving simultaneously developing surrounding tissues leads to regionalization in the eye anlage, namely the formation of the RPE and NR domains [18][55][56]. The major transcriptional modulators to differentiate and maintain the specific RPE properties are Mitf, Otx, and β-catenin. However, after the determination of the domains, both cell populations retain the ability to convert one into another (RPE⟷NR). In birds, this is observed during embryonic development [57], as well as in the case of BMP application [58]. The RPE conversion into the NR is also reported for amphibians [59][60] (see below). In Chx10-null mutant mice, NR cells transdifferentiate into RPE cells against the background of developing microphthalmia [61].
As mentioned above, during the NR development, RPCs produce the full range of retinal cell types. They do not do this simultaneously, although frequently with overlapping. This indicates that, already in the early phase of NR genesis, RPCs represent a heterogeneous population that exhibits differential gene expression responsible for differences in the RPC potential to produce one or another cell type [62].
One family of TFs that has been shown to be most important in the regulation of cell fate is the basic–loop–helix (bHLH) family. The proneural bHLH transcriptional regulators are key components for the intrinsic programming of RPCs and are essential for the formation of the diverse retinal cell types [63][64]. In turn, the expression of key intrinsic regulators, multiple retinogenic bHLH and homeodomain TFs, responsible for the specification of NR neurons, is controlled by the Pax6 master gene [65][66].
Miesfeld et al. [67], using antibodies recognizing the Atoh7 (formerly Math5) polypeptide of mice and humans, as well as informative knockout and transgenic mouse tissues and overexpression experiments, found that the transient features of the Atoh7 protein and Atoh7 mRNA expression during retinal neurogenesis match the expected pattern on the tissue and cellular levels during the neurogenesis wave preceding the optic cup formation. The differentiational expression of TFs Olig2, Ngn2 (Neurog2), and Ascl1 (Mash1) is characteristic of RPC subpopulations [68][69][70]. Brzezinski et al. [71] attempted to link the heterogeneity of the RPC population with the differences of its cells in the expression of TFs-encoding genes. They used genetically modified mouse lines, their embryos, as well as retinal explants in vitro. Ascl1- and Ngn2-inducible expression fate mapping was conducted using the CreER™/LoxP system. It was found that RPCs represent a highly heterogeneous cell population expressing every possible combination of TFs. By summarizing the results, it became possible to divide the RPC population into at least two categories: early (Ngn2+), which gives rise to a population of ganglion cells, and late (Ascl1+), whose cells differentiate into other types of retinal neurons. Simultaneously, it has been shown that the DNA-binding protein, Ikzf1/Ikaros zinc finger TF, plays a role in determining the state of RPCs’ competence associated with generating early-born cell types. The inactivation of Ikzf1 caused a loss of early-born neurons including ganglion, amacrine, and horizontal cells without affecting late-born cell types [72].
In the last 25 years, a large number of TFs, combinations of genes, as well as cofactors responsible for the differentiation of specific NR cell types have been identified using molecular genetics approaches and bioinformatics. In particular, it was found that Atoh7 and Pou4f2 (homeobox) catalyze the rate-limiting step in the specification of retinal ganglion cells. Prox1 expression proved to be essential for the production of horizontal cells, while Neurod1 and Neurod4 and Pax6 and Six3 regulate the production of amacrine cells. Crx is regarded as a key TF for the specification of photoreceptors, while Vsx2 (Chx10) is key for the production of bipolars [73][74]. The expression of TFs Sox11 and Sox4, carried out in a coordinated manner during retinogenesis, is responsible for correcting the size of populations of certain specific cell types. As reported, the epigenetic mechanisms of gene regulation are used in this case [75].
The scope of the present entry does not allow a detailed consideration of the dynamics of TFs’ expression during the maturation of each individual NR cell type. A more detailed description of the data found in this area of research is provided in earlier reviews, e.g., [14][19][76]. In many studies, a core transcriptional hierarchy underlying retinal cell types’ appearance in vertebrates is assumed. However, at the same time, in many studies, the complicated pattern of TFs’ expression is reported. It has been noted that TFs’ expression is influenced by simultaneous, synergistical expression of other TFs (combinatorial codes), which also plays an important role in cell fate diversification [77][78]. In RPCs, thousands of genes undergo differential expression, which are turned on or off as the major specific NR cell types arise. Although studies of the recent past have identified the key genetic regulators of retina specification, the question as to how their network works is still far from being fully addressed. In the review by Buono and Martinez-Morales [79], the authors confidently state that the system biology and the emerging techniques such as RNA-seq, ChIP-seq, ATAC-seq, or single-cell RNA-seq can make a significant contribution to understanding the operation principles of genetic regulatory networks in retinal development. Recently, Lyu et al. [76] used integrated single-cell RNA and single-cell ATAC sequencing (scATAC-seq) analysis and the models of developing mouse and human retinas to identify multiple interconnected, evolutionarily conserved genetic networks composed of cell-type-specific TFs that both activate genes within their own network and inhibit genes in other networks. It has been shown that such regulatory machinery can control temporal patterning in primary RPCs, regulate transition from primary to neurogenic progenitors, and drive the specification of each major retinal cell type. The authors exemplify this by TF nuclear factor I (NFI), which binds CAATT-boxes. It was indicated that this and other TFs selectively activate the expression of genes promoting late-stage temporal identity in primary retinal progenitors [76].
Along with studies of regulatory genetic networks, with the full complexity of this problem, attention is paid to epigenetic regulatory mechanisms, including the chromatin landscape, histone modifications, DNA methylation, non-encoded RNAs, etc. Thousands of enhancers are shown to be active in the developing retinae, and many of them have features of cell- and developmental stage-specific activity [80][81][82][83]. Attempts to identify the role of miRNAs were made earlier. MicroRNAs, single-stranded 19 to 25 nt small ncRNA that are part of the RNA-induced silencing complex (they pair with target sites located primarily within the 3′-untranslated region of mRNAs), are capable of suppressing gene expression by inhibiting the translation or causing degradation of RNA [84]. A study of the miRNAs profile by the application of the in situ hybridization method has shown that many of them are expressed both during retinogenesis and at the adult stage in overlapping and distinct patterns [85][86][87][88].
Norrie et al. [81] studied the dynamic changes in the 3D chromatin landscape by ultra-deep in situ Hi-C analysis on murine retinae during retinal development. Developmental stage-specific changes were identified in chromatin compartments and enhancer–promoter interactions. The authors designed a machine-learning-based algorithm to map euchromatin and heterochromatin domains’ genome-wide and overlaid it with chromatin compartments identified by Hi-C. Single-cell ATAC-seq and RNA-seq were integrated with Hi-C and previous ChIP-seq data to identify cell- and developmental-stage-specific super-enhancers. As a result, it became possible to identify the bipolar neuron-specific core regulatory circuit super-enhancers upstream of Vsx2, whose deletion in mice led to the loss of bipolar neurons [81]. A number of works consider the impact of the loss of polycomb repressive complex 2 (PRC2), which catalyzes the addition of the repressive mark H3K27me3, on retinal development. Mutations in the subunit of this complex (Ezh2, Ead) led to a noticeable decrease in the RPC proliferation, as well as to changes in the choice of the type of differentiation by progenitors [89][90].
Thus, studies considering the work of the genome and epigenome, the changes and modulations occurring in cells of the prospective retina as it self-organizes in vivo, being fundamental, are, nevertheless, still far from comprehensively addressing these issues. However, as can be seen, the prospects for such research have been outlined, and a technological capacity for complex molecular genetics and epigenetic studies has been built.
4. Morphogenetic Factors of Retinal Self-Organization In Vivo
The self-development of the retina implies the acquisition of its characteristic shape, the process of morphogenesis that occurs along with cell differentiation, and the formation of their coordinated functional circuit. In studying the role of TFs as morphogenetic factors, special attention is paid to Pax6, a master gene that is key to the control of eye development in both invertebrates and vertebrates [91][92]. In the work by Grocott et al. [93], who used chicks as a model organism, Pax6 was shown to direct the expression of a pair of morphogen coding genes, Fst and Tgfb2, which modulate the Pax6 function via positive and negative feedback. The topology of the Pax6/Fst/Tgfb2 gene network proved to be consistent with the activator–inhibitor-type Turing network [94], which is capable of manifesting a self-organizing pattern-forming ability in the absence of position information. This process was computationally modeled, and the results indicated that the work of this genetic network is essential for establishing the primary axis of organization for the spontaneously polarizing retina and prefiguring its further development [93].
Nevertheless, the study of the TF expression, signaling pathways, and their regulatory networks does not seem to be sufficient enough to explain the mechanisms of self-organization and morphogenesis of the retina. In particular, it is not clear how individual cells determine the state in space and time and consistently modulate the formation of a 3D structure until the definitive shape that suits the function to be performed. As discussed below, the extensive reshaping of the eye primordium during development is highly conserved across vertebrates and is successfully reproduced in vitro in organoid models, using not only RPCs, but also embryonic (ESCs) or induced pluripotent stem cells (iPSCs) [95][96]. This confirms the existence of the self-regulating properties of the RPCs and their ensembles involved in this process, which coordinate their actions by direct and indirect interactions between each other. The information about the molecular participants of these interactions in the early phases of self-organization is still insufficient [96].
Cellular mechanosensing mechanisms, which, apparently, should also be considered in the system of regulatory mechanisms of retinal morphogenesis, have been found using models of the development of other tissues [97][98]. The study of Okuda et al.[99] considers the mechanical aspect of the problem of eye and retina formation. The authors based their study on the previous data of experiments with the eye cups derived from mouse ESCs in vitro and the “relaxation–expansion” model proposed at that time [95][100]. In a study of cell displacements during optic cup formation, eye primordia of 9-day mouse embryos were used in ex vivo conditions. The key cell behaviors required for the invagination and the subsequent hinge formation along the NR–RPE boundary were identified. A conclusion was made that mechanical force plays a primary role in feeding back the 3D tissue deformation to the force generations of individual cells across different scales [99]. It is also known that mechanical deformations play a role in the formation of the optic fissure [101]. The Hippo signaling pathway, a main cell mechanotransducer that can respond to ECM stiffness, should also be mentioned in this context [102]. It is known that Yap and Taz, being the co-activators of the Hippo pathway, are involved in the RPE development as sensors of mechanical signals inside the cell nucleus [103]. Data obtained on a zebrafish model have shown that the Yap/Taz-Tead activity is necessary for a part of the RPCs, having equal potencies for both RPE and NR production in the optic vesicle, to acquire the RPE identity subsequently. The conclusion is supported by the Yap immunoreactivity in the nuclei of prospective RPE. Furthermore, zebrafish yap (yap1) mutants completely lost the population of RPE cells and/or exhibited NR colobomas (redundant proneural cell proliferation). These data allowed the conclusion that Yap and Taz are early key regulators of RPE genesis and one of the causes of congenital ocular defects [103]. The role of cell primary cilia, or rather the expression of genes regulating ciliogenesis, has recently been discovered in the morphogenesis of the eye. Fiora et al. [104] found that in Arl13-null mouse embryos, the lens is abnormally surrounded by an inverted optic cup whose RPE is oddly facing the surface ectoderm. It has been found also that Arl13b genes can modulate the work of the Shh signaling pathway along the dorsoventral (DV) axis and are thereby involved in setting the DV polarity and morphogenesis of the optic vesicle.
Many theoretical and practical attempts been made to show the role played by mechanics as a driver of tissue self-organization and morphogenesis [105]. Recently, attempts have been made to determine the role of RPE in the creation of the mechanical forces responsible for optic cup formation [106]. The authors managed to address this problem by genetically modifying the Tg (E1-bhlhe40:GFP) line of zebrafish. This allowed making all newly appearing RPE cells fluorescent. It was shown that, in the virtual absence of proliferation, RPE cells stretched and flattened, thereby matching the retinal curvature and promoting optic vesicle folding. The localized interference with the RPE cytoskeleton disrupted tissue stretching and optic vesicle folding.
The role of mechanical forces, as well as the ECM composition mentioned above, responsible for their generation, are traditionally discussed when considering the formation of the optic vesicle and the optic cup in vivo and in vitro, a well as in the development of congenital eye pathologies [107][108][109]. The ECM is involved in the regulation of the movements of individual cells and their groups; the binding of the ECM to the cell causes the contractile force to strengthen, with the subsequent transmission of this signal into the cell [107][110]. To describe the regulation of eye morphogenesis, when considering the role of mechanical forces and the ECM, authors often use mathematical models [93][101][107][111][112], as well as observations of changes in morphogenetic movements during the selective elimination of certain ECM proteins and their receptors [113][114]. Intraocular pressure (IOP), created through the accumulation of the aqueous humor of the eye, should also be mentioned among the physical forces that are factors influencing the formation of eye tissues in vivo. It has long been known that a slowdown/underdevelopment of the eye occurs with a decrease in IOP [115]. However, to date, there is no specific data on the role of flows of accumulated aqueous humor, the pressure they exert, and the resulting stresses in the tissue of the developing retina.
Proteins providing the planar cell polarity (PCP), i.e., a polarity axis that organizes cells in the plane of the tissue, are considered another morphogenetic regulatory factor. PCP proteins coordinate the planar polarity between cells and control polarized behavior by modulating the cytoskeleton [116]. According to Álvarez-Hernán et al. [117], the distribution of PCP proteins in the developing chick retina indicates their important role in the axonal guidance at early stages of retinogenesis, as well as possible involvement in the formation of cell asymmetry and the maintenance of retinal cell phenotypes.
Thus, when considering the phenomenon of retinal self-organization during eye development in vivo, the multifactorial nature of the process, including both intrinsic and extrinsic regulatory mechanisms, should be taken into account. In brief, these are as follows: cell–cell relationships and intercellular signaling positive and negative feedback, expression of genes and TFs and their networks operating differentially in a spatiotemporal manner, the role of epigenetic regulatory mechanisms, regulation carried out by external signaling pathways, cell proliferation, migration, and apoptosis, changes in mechanical properties and cell-level forces that are regulated by molecular signals, etc. This list is a vivid demonstration of the biological complexity and genetic heterogeneity of the self-organisation process.
References
- Heavner, W.; Pevny, L. Eye development and retinogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008391.
- Graw, J. Eye development. Curr. Top. Dev. Biol. 2010, 90, 343–386.
- Picker, A.; Cavodeassi, F.; Machate, A.; Bernauer, S.; Hans, S.; Abe, G.; Kawakami, K.; Wilson, S.W.; Brand, M. Dynamic coupling of pattern formation and morphogenesis in the developing vertebrate retina. PLoS Biol. 2009, 7, e1000214.
- Kwan, K.M.; Otsuna, H.; Kidokoro, H.; Carney, K.R.; Saijoh, Y.; Chien, C.-B. A complex choreography of cell movements shapes the vertebrate eye. Development 2012, 139, 359–372.
- Heermann, S.; Schütz, L.; Lemke, S.; Krieglstein, K.; Wittbrodt, J. Eye morphogenesis driven by epithelial flow into the optic cup facilitated by modulation of bone morphogenetic protein. eLife 2015, 4, e05216.
- Turner, D.L.; Cepko, C.L. A common progenitor for neurons and glia persists in rat retina late in development. Nature 1987, 328, 131–136.
- Wetts, R.; Fraser, S.E. Multipotent precursors can give rise to all major cell types of the frog retina. Science 1988, 239, 1142–1145.
- Holt, C.E.; Bertsch, T.W.; Ellis, H.M.; Harris, W.A. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron 1988, 1, 15–26.
- Cepko, C.L.; Austin, C.P.; Yang, X.; Alexiades, M.; Ezzeddine, D. Cell fate determination in the vertebrate retina. Proc. Natl. Acad. Sci. USA 1996, 93, 589–595.
- Harris, W.A. Cellular diversification in the vertebrate retina. Curr. Opin. Genet. Dev. 1997, 7, 651–658.
- Belliveau, M.J.; Cepko, C.L. Extrinsic and intrinsic factors control the genesis of amacrine and cone cells in the rat retina. Development 1999, 126, 555–566.
- Livesey, F.J.; Cepko, C.L. Vertebrate neural cell-fate determination: Lessons from the retina. Nat. Rev. Neurosci. 2001, 2, 109–118.
- Hoshino, A.; Ratnapriya, R.; Brooks, M.J.; Chaitankar, V.; Wilken, M.S.; Zhang, C.; Starostik, M.R.; Gieser, L.; La Torre, A.; Nishio, M.; et al. Molecular Anatomy of the Developing Human Retina. Dev. Cell. 2017, 43, 763–779.e4.
- Xiang, M. Intrinsic control of mammalian retinogenesis. Cell Mol. Life Sci. 2013, 70, 2519–2532.
- Bassett, E.A.; Wallace, V.A. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012, 35, 565–573.
- Akagi, T.; Inoue, T.; Miyoshi, G.; Bessho, Y.; Takahashi, M.; Lee, J.E.; Guillemot, F.; Kageyama, R. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J. Biol. Chem. 2004, 279, 28492–28498.
- Kim, H.-T.; Kim, J.W. Compartmentalization of Vertebrate Optic Neuroepithelium: External Cues and Transcription Factors. Mol. Cells 2012, 33, 317–324.
- Fuhrmann, S.; Zou, C.; Levine, E.M. Retinal pigment epithelium development, plasticity, and tissue homeostasis. Exp. Eye Res. 2014, 123, 141–150.
- Miesfeld, J.B.; Brown, N.L. Eye organogenesis: A hierarchical view of ocular development. Curr. Top. Dev. Biol. 2019, 132, 351–393.
- Boije, H.; MacDonald, R.B.; Harris, W.A. Reconciling competence and transcriptional hierarchies with stochasticity in retinal lineages. Curr. Opin. Neurobiol. 2014, 27, 68–74.
- Gomes, F.L.; Zhang, G.; Carbonell, F.; Correa, J.A.; Harris, W.A.; Simons, B.D.; Cayouette, M. Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development 2011, 138, 227–235.
- He, J.; Zhang, G.; Almeida, A.D.; Cayouette, M.; Simons, B.D.; Harris, W.A. How variable clones build an invariant retina. Neuron 2012, 75, 786–798.
- Green, E.S.; Stubbs, J.L.; Levine, E.M. Genetic rescue of cell number in a mouse model of microphthalmia: Interactions between Chx10 and G1-phase cell cycle regulators. Development 2003, 130, 539–552.
- Randlett, O.; Macdonald, R.B.; Yoshimatsu, T.; Almeida, A.D.; Suzuki, S.C.; Wong, R.O.; Harris, W.A. Cellular requirements for building a retinal neuropil. Cell Rep. 2013, 3, 282–290.
- Eldred, M.K.; Charlton-Perkins, M.; Muresan, L.; Harris, W.A. Self-organising aggregates of zebrafish retinal cells for investigating mechanisms of neural lamination. Development 2017, 144, 1097–1106.
- Trimarchi, J.M.; Stadler, M.B.; Cepko, C.L. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS ONE 2008, 3, e1588.
- Zakeri, Z.; Penaloza, C.G.; Smith, K.; Ye, Y.; Lockshin, R.A. What cell death does in development. Int. J. Dev. Biol. 2015, 59, 11–22.
- Vecino, E.; Acera, A. Development and programed cell death in the mammalian eye. Int. J. Dev. Biol. 2015, 59, 63–71.
- Valenciano, A.I.; Boya, P.; De La Rosa, E.J. Early neural cell death: Numbers and cues from the developing neuroretina. Int. J. Dev. Biol. 2009, 53, 1515–1528.
- Vecino, E.; Hernandez, M.; Garcia, M. Cell death in the developing vertebrate retina. Int. J. Dev. Biol. 2004, 48, 965–974.
- Braunger, B.M.; Demmer, C.; Tamm, E.R. Programmed cell death during retinal development of the mouse eye. Adv. Exp. Med. Biol. 2014, 801, 9–13.
- Fricker, M.; Tolkovsky, A.M.; Borutaite, V.; Coleman, M.; Brown, G.C. Neuronal Cell Death. Physiol. Rev. 2018, 98, 813–880.
- Shi, W.; Wang, R.; Niu, S.; Li, Y.; Ma, C.; Zhang, G.; Cong, B. Dynamic changes of proliferation and apoptosis in rat retina development. Int. J. Clin. Exp. Path. 2017, 10, 11679–11684.
- Amini, R.; Rocha-Martins, M.; Norden, C. Neuronal Migration and Lamination in the Vertebrate Retina. Front. Neurosci. 2018, 11, 742.
- Hoon, M.; Okawa, H.; Della Santina, L.; Wong, R.O. Functional architecture of the retina: Development and disease. Prog. Retin. Eye Res. 2014, 42, 44–84.
- Reese, B.E.; Keeley, P.W. Design principles and developmental mechanisms underlying retinal mosaics. Biol. Rev. Camb. Philos. Soc. 2015, 90, 854–876.
- Reese, B.E.; Harvey, A.R.; Tan, S.S. Radial and tangential dispersion patterns in the mouse retina are cell-class specific. Proc. Natl. Acad. Sci. USA 1995, 92, 2494–2498.
- Reese, B.E.; Necessary, B.D.; Tam, P.P.; Faulkner-Jones, B.; Tan, S.S. Clonal expansion and cell dispersion in the developing mouse retina. Eur. J. Neurosci. 1999, 11, 2965–2978.
- Kwan, K.M. Coming into focus: The role of extracellular matrix in vertebrate optic cup morphogenesis. Dev. Dyn. 2014, 243, 1242–1248.
- Fuhrmann, S.; Levine, E.M.; Reh, T.A. Extraocular mesenchyme patterns the optic vesicle during early eye development in the embryonic chick. Development 2000, 127, 4599–4609.
- Bryan, C.D.; Casey, M.A.; Pfeiffer, R.L.; Jones, B.W.; Kwan, K.M. Optic cup morphogenesis requires neural crest-mediated basement membrane assembly. Development 2020, 147, dev181420.
- Grocott, T.; Johnson, S.; Bailey, A.P.; Streit, A. Neural crest cells organize the eye via TGF-β and canonical Wnt signaling. Nat. Commun. 2011, 2, 266–269.
- Glinka, A.; Wu, W.; Onichtchouk, D.; Blumenstock, C.; Niehrs, C. Head induction by simultaneous repression of Bmp and Wnt signaling in Xenopus. Nature 1997, 389, 517–519.
- Lamba, D.A.; Karl, M.O.; Ware, C.B.; Reh, T.A. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2006, 103, 12769–12774.
- Pandit, T.; Jidigam, V.K.; Patthey, C.; Gunhaga, L. Neural retina identity is specified by lens-derived BMP signals. Development 2015, 142, 1850–1859.
- Pera, E.M.; Wessely, O.; Li, S.-Y.; De Robertis, E.M. Neural and head induction by insulin-like growth factor signals. Dev. Cell 2001, 1, 655–665.
- Richard-Parpaillon, L.; Heligon, C.; Chesnel, F.; Boujard, D.; Philpott, A. The IGF pathway regulates head formation by inhibiting Wnt signaling in Xenopus. Dev. Biol. 2002, 244, 407–417.
- Cavodeassi, F.; Carreira-Barbosa, F.; Young, R.M.; Concha, M.L.; Allende, M.L.; Houart, C.; Tada, M.; Wilson, S.W. Early Stages of Zebrafish Eye Formation Require the Coordinated Activity of Wnt11, Fz5, and the Wnt/β-Catenin Pathway. Neuron 2005, 47, 43–56.
- Fujimura, N. WNT/β-Catenin Signaling in Vertebrate Eye Development. Front. Cell Dev. Biol. 2016, 4, 138.
- Martínez-Morales, J.R.; Rodrigo, I.; Bovolenta, P. Eye development: A view from the retina pigmented epithelium. Bioessays 2004, 26, 766–777.
- Wagstaff, P.E.; Berzal, A.H.; Boon, C.J.F.; Quinn, P.M.J.; ten Asbroek, A.L.M.A.; Bergen, A.A. The Role of Small Molecules and Their Effect on the Molecular Mechanisms of Early Retinal Organoid Development. Int. J. Mol. Sci. 2021, 22, 7081.
- Balasubramanian, R.; Min, X.; Quinn, P.M.J.; Giudice, Q.L.; Tao, C.; Polanco, K.; Makrides, N.; Peregrin, J.; Bouaziz, M.; Mao, Y.; et al. Phase transition specified by a binary code patterns the vertebrate eye cup. Sci. Adv. 2021, 7, eabj9846.
- Seritrakul, P.; Gross, J.M. Genetic and epigenetic control of retinal development in zebrafish. Curr. Opin. Neurobiol. 2019, 59, 120–127.
- Kenyon, K.L.; Zaghloul, N.; Moody, S.A. Transcription factors of the anterior neural plate alter cell movements of epidermal progenitors to specify a retinal fate. Dev. Biol. 2001, 240, 77–91.
- Sinn, R.; Wittbrodt, J. An eye on eye development. Mech. Dev. 2013, 130, 347–358.
- Zuber, M.E.; Gestri, G.; Viscian, A.S.; Barsacchi, G.; Harris, W.A. Specification of the vertebrate eye by a network of eye field transcription factors. Development 2003, 130, 5155–5167.
- Luz-Madrigal, A.; Grajales-Esquivel, E.; McCorkle, A.; DiLorenzo, A.M.; Barbosa-Sabanero, K.; Tsonis, P.A.; Del Rio-Tsonis, K. Reprogramming of the chick retinal pigmented epithelium after retinal injury. BMC Biol. 2014, 12, 28.
- Steinfeld, J.; Steinfeld, I.; Bausch, A.; Coronato, N.; Hampel, M.-L.; Depner, D.; Layer, P.G.; Vogel-Höpker, A. BMP-induced reprogramming of the neural retina into retinal pigment epithelium requires Wnt signaling. Biol. Open. 2017, 6, 979–992.
- Mitashov, V.I. Mechanisms of retina regeneration in vertebrates. Int. J. Dev. Biol. 1996, 40, 833–844.
- Yoshii, C.; Ueda, Y.; Okamoto, M.; Araki, M. Neural retinal regeneration in the anuran amphibian Xenopus laevis post-metamorphosis: Transdifferentiation of retinal pigmented epithelium regenerates the neural retina. Dev. Biol. 2007, 303, 45–50.
- Rowan, S.; Chen, C.M.; Young, T.L.; Fisher, D.E.; Cepko, C.L. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development 2004, 131, 5139–5152.
- Watanabe, T.; Raff, M.C. Rod photoreceptor development in vitro: Intrinsic properties of proliferating neuroepithelial cells change as development proceeds in the rat retina. Neuron 1990, 4, 461–467.
- Hatakeyama, J.; Kageyama, R. Retinal cell fate determination and bHLH factors. Semin. Cell Dev. Biol. 2004, 15, 83–89.
- Mao, C.A.; Cho, J.H.; Wang, J.; Gao, Z.; Pan, P.; Tsai, W.W.; Frishman, L.J.; Klein, W.H. Reprogramming amacrine and photoreceptor progenitors into retinal ganglion cells by replacing Neurod1 with Atoh7. Development 2013, 140, 541–551.
- Marquardt, T.; Ashery-Padan, R.; Andrejewski, N.; Scardigli, R.; Guillemot, F.; Gruss, P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell 2001, 105, 43–55.
- Oron-Karni, V.; Farhy, C.; Elgart, M.; Marquardt, T.; Remizova, L.; Yaron, O.; Xie, Q.; Cvekl, A.; Ashery- Padan, R. Dual requirement for Pax6 in retinal progenitor cells. Development 2008, 135, 4037–4047.
- Miesfeld, J.B.; Glaser, T.; Brown, N.L. The dynamics of native Atoh7 protein expression during mouse retinal histogenesis, revealed with a new antibody. Gene Expr. Patterns 2018, 27, 114–121.
- Jasoni, C.L.; Reh, T.A. Temporal and spatial pattern of MASH-1 expression in the developing rat retina demonstrates progenitor cell heterogeneity. J. Comp. Neurol. 1996, 369, 319–327.
- Nakamura, K.; Harada, C.; Namekata, K.; Harada, T. Expression of olig2 in retinal progenitor cells. NeuroReport 2006, 17, 345–349.
- Shibasaki, K.; Takebayashi, H.; Ikenaka, K.; Feng, L.; Gan, L. Expression of the basic helix-loop-factor Olig2 in the developing retina: Olig2 as a new marker for retinal progenitors and late-born cells. Gene Expr. Patterns 2007, 7, 57–65.
- Brzezinski, J.A., 4th; Kim, E.J.; Johnson, J.E.; Reh, T.A. Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development 2011, 138, 3519–3531.
- Elliott, J.; Jolicoeur, C.; Ramamurthy, V.; Cayouette, M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron 2008, 60, 26–39.
- Ohsawa, R.; Kageyama, R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Res. 2008, 1192, 90–98.
- Clark, A.M.; Yun, S.; Veien, E.S.; Wu, Y.Y.; Chow, R.L.; Dorsky, R.I.; Levine, E.M. Negative regulation of Vsx1 by its paralog Chx10/Vsx2 is conserved in the vertebrate retina. Brain Res. 2008, 1192, 99–113.
- Usui, A.; Mochizuki, Y.; Iida, A.; Miyauchi, E.; Satoh, S.; Sock, E.; Nakauchi, H.; Aburatani, H.; Murakami, A.; Wegner, M.; et al. The early retinal progenitor-expressed gene Sox11 regulates the timing of the differentiation of retinal cells. Development 2013, 140, 740–750.
- Lyu, P.; Hoang, T.; Santiago, C.P.; Thomas, E.D.; Timms, A.E.; Appel, H.; Gimmen, M.; Le, N.; Jiang, L.; Kim, D.W.; et al. Gene regulatory networks controlling temporal patterning, neurogenesis, and cell-fate specification in mammalian retina. Cell Rep. 2021, 37, 109994.
- Wang, J.C.C.; Harris, W.A. The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev. Biol. 2005, 285, 101–115.
- Zhang, X.; Serb, J.M.; Greenlee, M.H. Mouse retinal development: A dark horse model for systems biology research. Bioinform. Biol. Insights 2011, 5, 99–113.
- Buono, L.; Martinez-Morales, J.R. Retina development in vertebrates: Systems biology approaches to understanding genetic programs: On the contribution of next-generation sequencing methods to the characterization of the regulatory networks controlling vertebrate eye development. Bioessays 2020, 42, e1900187.
- Aldiri, I.; Xu, B.; Wang, L.; Chen, X.; Hiler, D.; Griffiths, L.; Valentine, M.; Shirinifard, A.; Thiagarajan, S.; Sablauer, A.; et al. The Dynamic Epigenetic Landscape of the Retina During Development, Reprogramming, and Tumorigenesis. Neuron 2017, 94, 550–568.e10.
- Norrie, J.L.; Lupo, M.S.; Xu, B.; Al Diri, I.; Valentine, M.; Putnam, D.; Griffiths, L.; Zhang, J.; Johnson, D.; Easton, J.; et al. Nucleome Dynamics during Retinal Development. Neuron 2019, 104, 512–528.e11.
- Raeisossadati, R.; Ferrari, M.F.R.; Kihara, A.H.; AlDiri, I.; Gross, J.M. Epigenetic regulation of retinal development. Epigenetics Chromatin 2021, 14, 11.
- Daghsni, M.; Aldiri, I. Building a Mammalian Retina: An Eye on Chromatin Structure. Front. Genet. 2021, 12, 775205.
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531.
- Xu, S.; Witmer, P.D.; Lumayag, S.; Kovacs, B.; Valle, D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 2007, 282, 25053–25066.
- Ryan, D.G.; Oliveira-Fernandes, M.; Lavker, R.M. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol. Vis. 2006, 12, 1175–1184.
- Hackler, L., Jr.; Wan, J.; Swaroop, A.; Qian, J.; Zack, D.J. MicroRNA profile of the developing mouse retina. Investig. Ophthalmol. Vis. Sci. 2010, 51, 1823–1831.
- Cremisi, F. MicroRNAs and cell fate in cortical and retinal development. Front. Cell Neurosci. 2013, 7, 141.
- Zhang, J.; Taylor, R.J.; La Torre, A.; Wilken, M.S.; Cox, K.E.; Reh, T.A.; Vetter, M.L. Ezh2 Maintains Retinal Progenitor Proliferation, Transcriptional Integrity, and the Timing of Late Differentiation. Dev. Biol. 2015, 403, 128–138.
- Fujimura, N.; Kuzelova, A.; Ebert, A.; Strnad, H.; Lachova, J.; Machon, O.; Busslinger, M.; Kozmik, Z. Polycomb Repression Complex 2 Is Required for the Maintenance of Retinal Progenitor Cells and Balanced Retinal Differentiation. Dev. Biol. 2018, 433, 47–60.
- Chow, R.L.; Altmann, C.R.; Lang, R.A.; Hemmati-Brivanlou, A. Pax6 induces ectopic eyes in a vertebrate. Development 1999, 126, 4213–4222.
- Gehring, W.J. Chance and Necessity in Eye Evolution. Genome Biol. Evol. 2011, 3, 1053–1066.
- Grocott, T.; Lozano-Velasco, E.; Mok, J.F.; Münsterberg, A.E. The Pax6 master control gene initiates spontaneous retinal development via a self-organising Turing network. Development 2020, 147, dev185827.
- Turing, A. The Chemical Basis of Morphogenesis (PDF). Philos. Trans. R. Soc. Lond. B 1952, 237, 37–72.
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56.
- Nakano, T.; Ando, S.; Takata, N.; Kawada, M.; Muguruma, K.; Sekiguchi, K.; Saito, K.; Yonemura, S.; Eiraku, M.; Sasai, Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012, 10, 771–785.
- D’Souza, S.; Lang, L.A. Retinal ganglion cell interactions shape the developing mammalian visual system. Development 2020, 147, dev196535.
- Discher, D.E.; Janmey, P.; Wang, Y.-L. Tissue cells feel and respond to the stiffness of their substrate. Science 2005, 310, 1139–1143.
- Polacheck, W.J.; German, A.E.; Mammoto, A.; Ingber, D.E.; Kamm, R.D. Mechanotransduction of fluid stresses governs 3D cell migration. Proc. Natl. Acad. Sci. USA 2014, 111, 2447–2452.
- Okuda, S.; Takata, N.; Hasegawa, Y.; Kawada, M.; Inoue, Y.; Adachi, T.; Sasai, Y.; Eiraku, M. Strain-triggered mechanical feedback in self-organizing optic-cup morphogenesis. Sci. Adv. 2018, 4, eaau1354.
- Eiraku, M.; Adachi, N.; Sasai, Y. Relaxation-expansion model for self-driven retinal morphogenesis: A hypothesis from the perspective of biosystems dynamics at the multi-cellular level. Bioessays 2012, 34, 17–25.
- Ching Chan, B.H.; Moosajee, M.; Rainger, J. Closing the Gap: Mechanisms of Epithelial Fusion during Optic Fissure Closure. Front. Cell Dev. Biol. 2020, 8, 620774.
- Chakraborty, S.; Njah, K.; Pobbati, A.V.; Lim, Y.B.; Raju, A.; Lakshmanan, M.; Tergaonkar, V.; Lim, C.T.; Hong, W. Agrin as a mechanotransduction signal regulating YAP through the Hippo pathway. Cell Rep. 2017, 18, 2464–2479.
- Miesfeld, J.B.; Gestri, G.; Clark, B.S.; Flinn, M.A.; Poole, R.J.; Bader, J.R.; Besharse, J.C.; Wilson, S.W.; Link, B.A. Yap and Taz regulate retinal pigment epithelial cell fate. Development 2015, 142, 3021–3032.
- Gomez-Galvez, P.; Anbari, S.; Escudero, L.M.; Buceta, J. Mechanics and self-organization in tissue development. Semin. Cell Dev. Biol. 2021, 120, 147–159.
- Moreno-Mármol, T.; Ledesma-Terrón, M.; Tabanera, N.; Martin-Bermejo, M.J.; Cardozo, J.M.J.; Cavodeassi, F.; Bovolenta, P. Stretching of the retinal pigment epithelium contributes to zebrafish optic cup morphogenesis. eLife 2021, 10, e63396.
- Hosseini, H.S.; Taber, L.A. How mechanical forces shape the developing eye. Prog. Biophys. Mol. Biol. 2018, 137, 25–36.
- Cavodeassi, F. Dynamic Tissue Rearrangements during Vertebrate Eye Morphogenesis: Insights from Fish Models. J. Dev. Biol. 2018, 6, 4.
- Fortune, B. Pulling and Tugging on the Retina: Mechanical Impact of Glaucoma Beyond the Optic Nerve Head. Investig. Ophthalmol. Vis. Sci. 2019, 60, 26–35.
- Nicolás-Pérez, M.; Kuchling, F.; Letelier, J.; Polvillo, R.; Wittbrodt, J.; Martínez-Morales, J.R. Analysis of cellular behavior and cytoskeletal dynamics reveal a constriction mechanism driving optic cup morphogenesis. eLife 2016, 5, e15797.
- Eiraku, M.; Sasai, Y. Self-formation of layered neural structures in three-dimensional culture of ES cells. Curr. Opin. Neurobiol. 2012, 22, 768–777.
- Roberts, P.A.; Gaffney, E.A.; Luthert, P.J.; Foss, A.J.E.; Byrne, H.M. Mathematical and computational models of the retina in health, development and disease. Prog. Retin. Eye Res. 2016, 53, 48–69.
- Oltean, A.; Huang, J.; Beebe, D.C.; Taber, L.A. Tissue growth constrained by extracellular matrix drives invagination during optic cup morphogenesis. Biomech. Model. Mechanobiol. 2016, 15, 1405–1421.
- Clements, R.; Turk, R.; Campbell, K.P.; Wright, K.M. Dystroglycan Maintains Inner Limiting Membrane Integrity to Coordinate Retinal Development. J. Neurosci. 2017, 37, 8559–8574.
- Coulombre, A. The role of intraocular pressure in the development of the chick eye. J. Exp. Zool. 1956, 133, 211–225.
- Butler, M.T.; Wallingford, J.B. Planar cell polarity in development and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 375–388.
- Álvarez-Hernán, G.; Garrido-Jiménez, S.; Román, A.C.; Carvajal-González, H.M.; Francisco-Morcillo, J. Distribution of planar cell polarity proteins in the developing avian retina. Exp. Eye Res. 2021, 209, 108681.
More
Information
Subjects:
Developmental Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.5K
Revisions:
2 times
(View History)
Update Date:
30 Jun 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





