Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Yun Kong | -- | 2148 | 2022-06-20 07:21:16 | | | |
| 2 | Sirius Huang | Meta information modification | 2148 | 2022-06-20 11:31:25 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kong, Y.; Wang, Y.; Miao, L.; Mo, S.; Li, J.; Zheng, X. Anticyanobacterial Modes and Mechanisms against Microcystis aeruginosa. Encyclopedia. Available online: https://encyclopedia.pub/entry/24200 (accessed on 07 February 2026).
Kong Y, Wang Y, Miao L, Mo S, Li J, Zheng X. Anticyanobacterial Modes and Mechanisms against Microcystis aeruginosa. Encyclopedia. Available at: https://encyclopedia.pub/entry/24200. Accessed February 07, 2026.
Kong, Yun, Yue Wang, Lihong Miao, Shuhong Mo, Jiake Li, Xing Zheng. "Anticyanobacterial Modes and Mechanisms against Microcystis aeruginosa" Encyclopedia, https://encyclopedia.pub/entry/24200 (accessed February 07, 2026).
Kong, Y., Wang, Y., Miao, L., Mo, S., Li, J., & Zheng, X. (2022, June 20). Anticyanobacterial Modes and Mechanisms against Microcystis aeruginosa. In Encyclopedia. https://encyclopedia.pub/entry/24200
Kong, Yun, et al. "Anticyanobacterial Modes and Mechanisms against Microcystis aeruginosa." Encyclopedia. Web. 20 June, 2022.
Copy Citation
Harmful algal blooms (HABs) have attracted great attention around the world due to the numerous negative effects such as algal organic matters and cyanobacterial toxins in drinking water treatments. Among the blooming cyanobacteria, Microcystis aeruginosa is one of the most common and widespread species. As an economic and environmentally friendly technology, microorganisms have been widely used for pollution control and remediation, especially in the inhibition/biodegradation of the toxic cyanobacterium Microcystis aeruginosa in eutrophic water; moreover, some certain anticyanobacterial microorganisms can degrade microcystins at the same time.
Microcystis aeruginosa
microorganisms
biodegradation
anticyanobacterial modes
harmful cyanobacterial blooms
1. Introduction
Harmful cyanobacterial blooms (HCBs) caused by cyanobacteria (including Microcystis, Anabaena, Nodularia, Oscillatoria, and so on) have become a common occurrence in freshwater worldwide [1][2]. Among the blooming cyanobacteria, Microcystis aeruginosa is one of the most common and widespread species [3]; specifically, it is known to be a representative species due to the dominant production of microcystins [4][5]. The rapid and excessive growth of M. aeruginosa is harmful to drinking water treatments and aquatic ecosystems due to the release of algal organic matters and cyanobacterial toxins [6][7]. As a result, the control of HCBs in water sources is a matter of great urgency.
Many approaches have been adopted for M. aeruginosa removal over the past few decades [8]. Physical methods including mechanical salvage, physical aeration, and ultrasonic treatment are usually high cost and take a long time; chemical methods such as chemical oxidants are highly efficient and low-cost methods for controlling HCBs within a short time [9]. However, chemicals may lead to a secondary contamination that may lead to potential threats to the aquatic ecosystem [10][11]. Compared with the physical and chemical methods, biological approaches such as plant allelopathy, aquatic animals and anticyanobacterial microorganisms are considered to be an economic and environmentally friendly way for cyanobacteria inhibition/biodegradation [2][10][12]. Among these methods, anticyanobacterial microorganisms are used as efficient biological agents M. aeruginosa [13]; furthermore, the microcystins can be biodegraded by certain anticyanobacterial microorganisms at the same time [6][14][15].
2. Anticyanobacterial Modes
In general, the anticyanobacterial modes by microorganisms are divided into direct attack (bacterial and cyanobacterial cell contact) and indirect attack (the release of anticyanobacterial substances) (Figure 1) [10][16][17][18]. To date, although anticyanobacteria can directly kill several different kinds of cyanobacteria, only few has been reported. A wide range of cyanobacteria including M. aeruginosa, M. wesenbergii, M. viridis, Anabaena flos-aquae, Oscillatoria tenuis, Nostoc punctiforme and Spirulina maxima are lysed by B. cereus DC22 with the direct attack mode, as well as chlorophyceae (Chlorella ellipsoidea and Selenastrum capricornutum) [19]. In addition to B. cereus, other anticyanobacteria that destroy M. aeruginosa with direct attack have also been reported. For example, the anticyanobacterial modes of Aeromonas bestiarum HYD0802-MK36 [20], Chryseobacterium sp. [21], Streptomyces globisporus G9 [22], Alcaligenes denitrificans [23], and Shigella sp. H3 [24] on M. aeruginosa are regarded as direct attack, and a number of cyst-like cells are formed in cyanobacteria during the direct attack [10]. It is speculated that the cyanobacterial cell walls are partially destroyed at the contact point with the anticyanobacteria, and the formation of cyst-like cells is a potential defense system against anticyanobacteria [2][10].
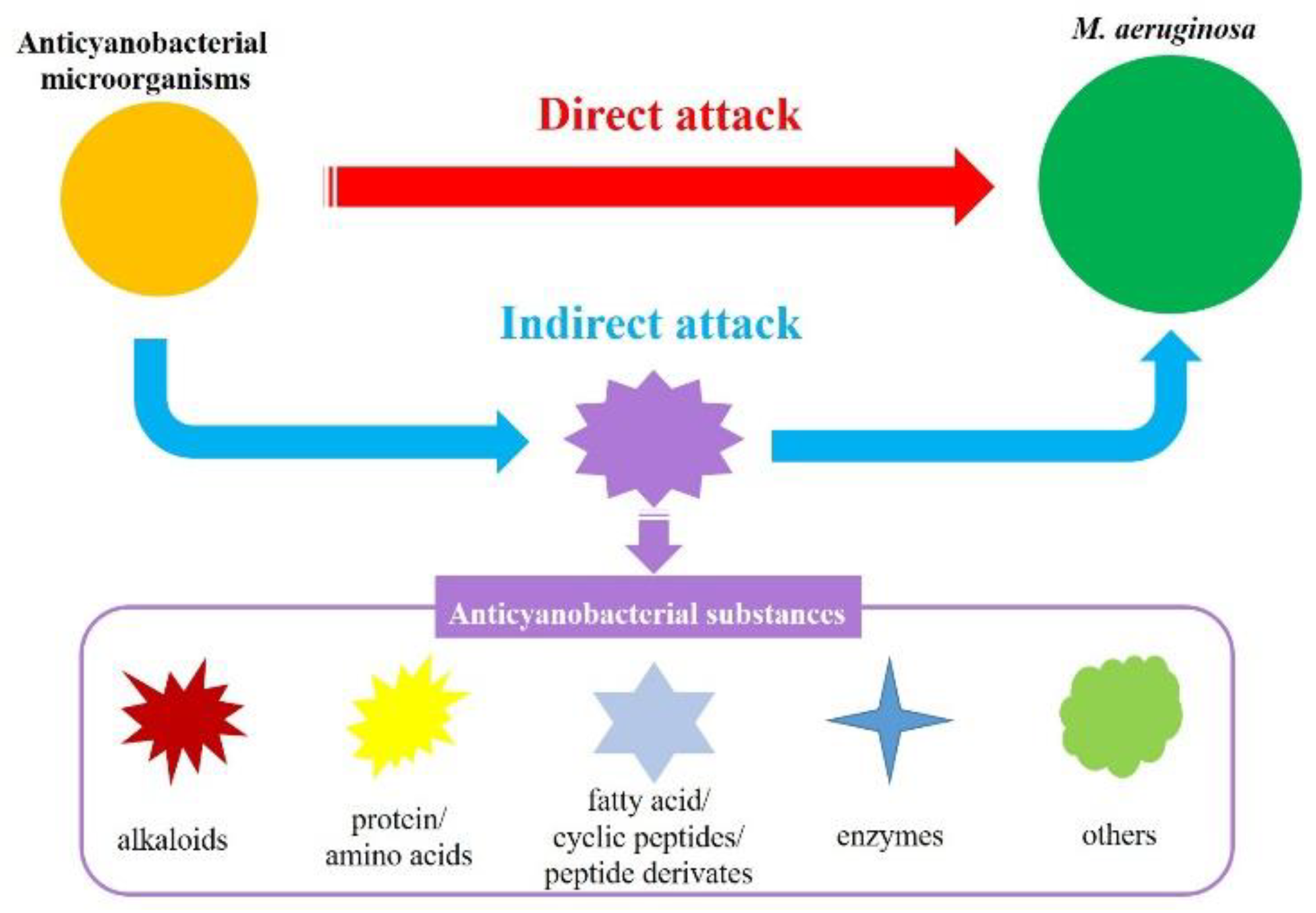
Figure 1. Anticyanobacterial modes of microorganisms against M. aeruginosa.
The indirect attack mode has been observed in the numerous metabolites from most of the reported anticyanobacterial microorganisms, and the anticyanobacterial characteristics of these bacteria seem to be unique to M. aeruginosa. Up to now, the genus Acinetobacter [17][25][26] and Exiguobacterium [27][28][29], which firstly attach to M. aeruginosa and then cause serious damage to the cyanobacterial cell structure and morphology, are recognized as degrading M. aeruginosa by producing anticyanobacterial substances. Nevertheless, some anticyanobacteria can inhibit or kill green alga and cyanobacteria with an indirect attack simultaneously. For instance, B. amyloliquefaciens FZB42 can efficiently eliminate M. aeruginosa, Anabaena sp., A. flos-aquae and Nostoc sp. by secreting bacilysin [30]. In line with this genus, B. amyloliquefaciens T1 produces amino acids to inhibit the growth of four Microcystis spp., but not of Anabaena flos-aquae or Chlorella pyrenoidosa [31][32]; S. amritsarensis HG-16 kills A. flos-aquae, Phormidium sp. and five Microcystis spp. by secreting active substances, but has a small inhibitory effect on C. vulgaris and a promoting effect on Oscillatoria sp. [5]. Along with this, the anticyanobacterial modes of Aquimarina salinaria on green algae and cyanobacterium, which is a direct attack on C. vulgaris 211-31 and an indirect attack on M. aeruginosa MTY01, is quite different [33]. Furthermore, a recent study firstly demonstrated that Paucibacter aquatile DH15 inhibits M. aeruginosa by both direct and indirect attacks [34], which would be interesting and could shed further light on the anticyanobacterial modes by microorganisms.
3. Anticyanobacterial Mechanisms
Currently, the anticyanobacterial mechanisms of microorganisms against M. aeruginosa are mainly dependeent on the attack modes, and these mechanisms are revealed with the changes in the photosynthesis system, antioxidant enzymes system, gene expression and QS system (Figure 2).
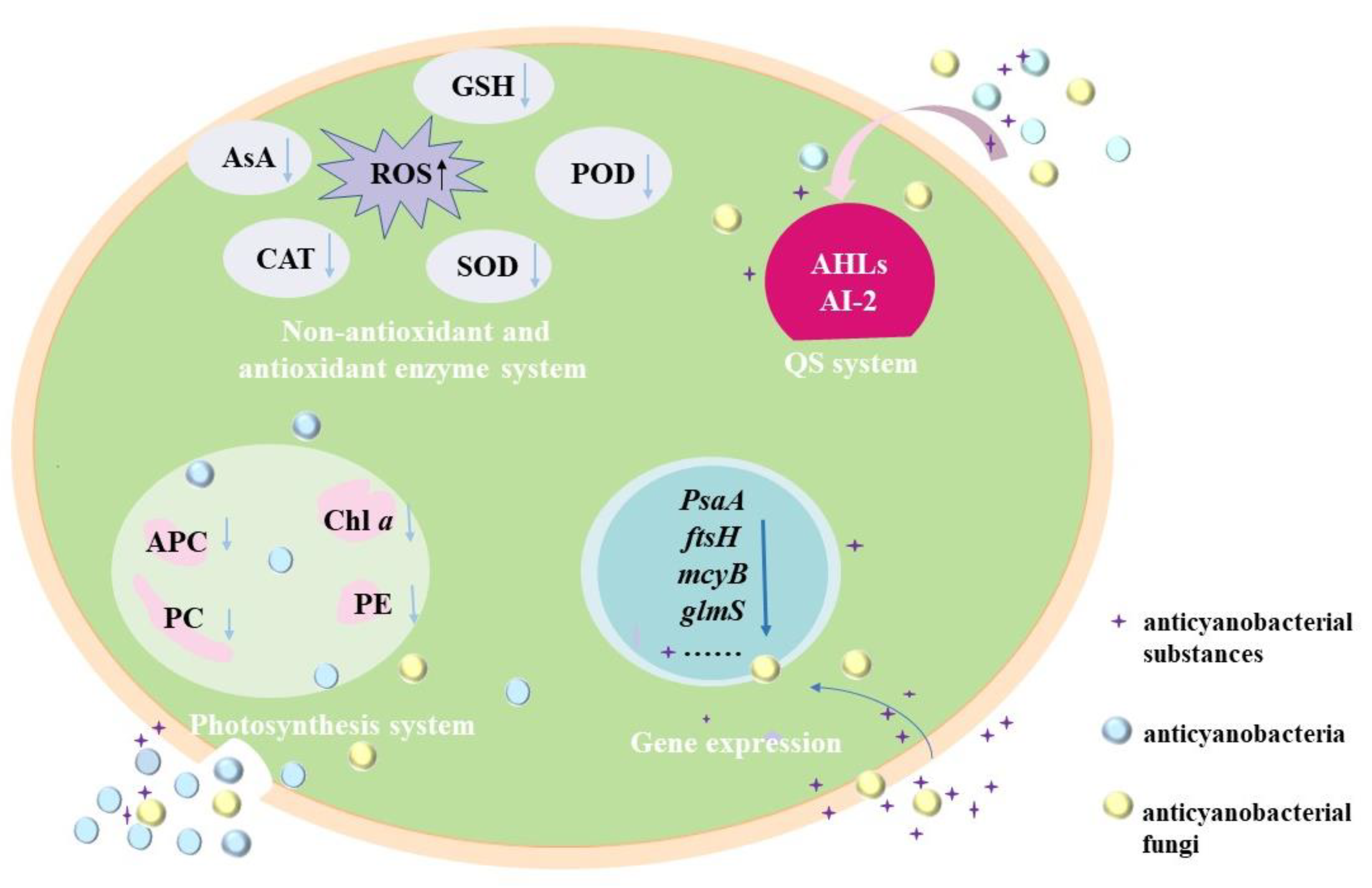
Figure 2. Anticyanobacterial mechanisms of microorganisms against M. aeruginosa.
3.1. Effects of Anticyanobacterial Microorganisms on Photosynthesis
Photosynthesis, which converts solar energy into chemical energy through the photosynthesis system (PS) II and PS I, is the principal mode of energy metabolism in cyanobacteria [35]. Anticyanobacterial microorganisms can significantly affect the photosynthesis of M. aeruginosa cells in several ways, including decreasing the chlorophyll a (Chl a) contents and photosynthetic pigments [36], and the disruption of the electron transport pathway in PS [37][38]. Chl a is one of the important components of cyanobacterial pigments. It is markedly decreased in M. aeruginosa under the exposure of anticyanobacteria such as P. aeruginosa [39][40], Streptomyces sp. [41][42], Exiguobacterium sp. [27][28], and so on. For the photosynthetic pigments, phycocyanobilin (PC), allophycocyanin (APC) and phycoerythrin (PE) are major indicators of cyanobacterial photosynthetic efficiency and are essential apparatus for light harvesting [34], and the addition of anticyanobacterium results in a significant decrease in the PC, APC and PE by disrupting the synthesis of an photosynthetic pigments [36]. In addition, the expressions of pcA and apcA genes for PC and APC synthesis in M. aeruginosa are down-regulated by Paucibacter aquatile DH15, which shows an inhibition effect on active chlorophyll [34]. It has been noted that the Chl a decrease is closely related to the reduction in photosynthetic pigments, and the cyanobacterial membrane is sensitive and easily damaged by anticyanobacterium [36].
The variations of cyanobacterial energy kinetics have also been evaluated by Chl fluorescence parameters, such as the maximum photochemical quantum yield of PS II (Fv/Fm), the effective quantum yield (Φe), and the maximum electron transport rate (ETRmax) [43][44]. With the addition of fermentation filtrate (5%, v/v) of Paenibacillus sp. SJ-73, the Fv/Fm values of M. aeruginosa PCC7806 and M. aeruginosa TH1701 dramatically decline from 0.52 and 0.29 to 0 [44]; similarly, it is only 0.08 (14.3% of the initial value) for M. aeruginosa FACHB-905 after being treated for 24 h by the fermentation filtrate (5%, v/v) of Raoultella sp. S1 [37]. Besides, the Φe and ETRmax of M. aeruginosa 9110 following the treatment of Chryseobacterium sp. GLY-1106 decrease gradually with time [43]; the ETRmax values of M. aeruginosa are also depressed significantly under the stress of Raoultella sp. S1 [37] and Bacillus sp. B50 [38]. The decreases in Fv/Fm, Φe and ETRmax demonstrate that the photosynthetic system is seriously damaged and the electron transport chain is blocked, resulting in the inhibition of cyanobacterial cell photosynthesis [45]. In consequence, the possible mechanism underlying the photosynthetic reduction could be due to the reduction in Fv/Fm, Φe and ETRmax in M. aeruginosa.
3.2. Effects of Anticyanobacterial Microorganisms on Antioxidant Enzymes System
The oxidative damage of the cyanobacterial cells can occur under different environmental stress conditions, and it will results in an increase in reactive oxygen species (ROS), which includes the superoxide anion radical, hydrogen peroxide and hydroxyl radicals [34][46]; while excess ROS often leads to oxidative stress, lipid peroxidation, and DNA damage [36][47]. The enzymatic antioxidants (such as catalase (CAT), superoxide dismutase (SOD), peroxidase (POD), and so on) and non-enzymatic antioxidants (such as ascorbic acid (AsA) and glutathione (GSH)) are responsible for removing the overproduction of ROS [2][43][48]. For instance, Streptomyces eurocidicus JXJ-0089 inhibits the growth of cyanobacterial cells in various ways, including promoting ROS production (e.g., O2•−), inhibiting the antioxidant synthesis, removing chlorophyll and destroying cell walls [49].
The ROS of cyanobacteria increases excessively by either the direct attack or indirect attack of anticyanobacterial microorganisms. The O2•− content in M. aeruginosa cells is induced largely by 4 μg mL−1 3, 4-dihydroxybenzalacetone (DBL) secreted from Phellinus noxius HN-1 and increased from 0.360 ± 0.001 to 0.400 ± 0.001 μg g−3 [50]. The ROS level of M. aeruginosa NIES 843 treated with Bacillus sp. AF-1 (cell-free filtrate) was lower than that of the control at the first 48 h but much higher at 72 h, indicating that some evasive mechanisms were taken to prevent the ROS accumulation in cyanobacterial cells at the initial stage [46]. Similar variations of ROS have been observed in M. aeruginosa KW after being treated with Paucibacter aquatile DH15, and the malondialdehyde (MDA) content and SOD activity related to remove ROS also increased at first and then decreased [34]; The MDA content, CAT and POD activity of M. aeruginosa FACHB-905 also increased quickly when fermentation liquid (5%, v/v) of P. aeruginosa [39] and P. chrysosporium was added quickly [51]; moreover, the responses of M. aeruginosa FACHB-905 cells to Streptomyces sp. KY-34 and Streptomyces sp. HJC-D1 following a similar pattern with the increases of CAT, SOD and POD, and the MDA further increased during the incubation time [36][47]. Although the antioxidants increased immediately to relieve the damage caused by anticyanobacteria, the cyanobacterial cell membrane may have decompose due to the accumulation of MDA [39][47][52].
For the non-enzymatic antioxidants, the variation of GSH is opposite to that of the antioxidase activity. The Bacillus licheniformis Sp34 induces more GSH production in M. aeruginosa at first to clear ROS, but the GSH content is much lower at 20 h (compared with the control) [53]. Such a phenomenon is also obtained in the anticyanobacterial process of Raoultella sp. S1 [37]. The prodigiosin from Hahella sp. KA22 also leads to the variation of GSH content, while the GSH content decreases slightly after exposure for 36 h [48]. These results demonstrate that the ROS levels and MDA contents decrease under prolonged exposure to anticyanobacteria [41][48][54]; in addition, the non-enzymatic antioxidants also play a critical role in protecting the cyanobacterial cells from oxidative damage under anticyanobacterial stress [37].
3.3. Effects of Anticyanobacterial Microorganisms on Gene Expression
The relative transcriptional level of some critical genes in cyanobacteria can be dramatically changed by anticyanobacterial microorganisms and substances, including genes related to the synthesis of photosystem reaction center proteins (PsaA, psaB, psbA1 and psbD1) [53][55], peptidoglycan synthesis (glmS), membrane proteins (ftsH), antioxidase (prx) [56], heat-shock proteins (grpE) [56], fatty acids (fabZ) [56], cyanotoxin microcystins (mcyA, mcyB, mcyC and mcyD) [22][57], the functions of cell division (ftsZ) [38], CO2 fixation (rbcL) [34], and DNA repair (ftsH and recA) [2][5]. Researchers have reported that the transcription expressions of genes ftsZ, psbA1, and glmS are decreased by DBL that is isolated from P. noxius HN-1 [50] and bacilysin that secreted from B. amyloliquefaciens FZB42 [30]. The expressions of gene ftsZ and psbA are also significantly inhibited by Bacillus sp. B50 [38], and the transcriptions of photosynthesis-related genes (psaB and psbD1) and CO2 fixation gene (rbcL) are inhibited by B. licheniformis Sp34 [53], indicating that the metabolisms of M. aeruginosa are destroyed. Other studies on transcriptomic analysis have demonstrated that the principal subunits of the reaction center (PsaA and PsaB) and other subunits (PsaC, PsaE, PsaD, PsaF and PsaL) are significantly down-regulated by B. laterosporus Bl-zj [55]. It is similar in the case of S. globisporus G9, S. amritsarensis and Raoultella sp. S1, which suppresses the expression of psbA1, psbD1 or rbcL [5][22][37]. The reduction in photosynthesis-related gene transcripts might result in an interruption in the electron transport chain and may finally affect the CO2 fixation process [34].
Gene such as mcyB that are involved in microcystins synthesis are also inhibited by Penicillium spp. [57], the white-rot fungi P. chrysosporium [51][56] and P. noxius HN-1 [50]; moreover, both directly attack the anticyanobacterium (S. globisporus G9) [22] and indirectly attack anticyanobacteria (including S. amritsarensis HG-16 and Bacillus sp. AF-1) could inhibit microcystins synthesis [5][46]. However, the inhibiting ability of Bacillus sp. AF-1 has not been confirmed with microcystins measurements [5].
3.4. Regulating the Anticyanobacterial Activity by QS System
QS system is the regulator control system for microorganisms that sense the cell density of their own species and make themselves to coordinate gene expression and physiological accommodation on a community scale [58][59]. It is a cell-to-cell communication that relies on the signal molecules [60], and the accumulated QS signals can bind to the cognate receptors and regulate biological activities and cellular functions [61][62]. Previous studies have shown that microbial behaviors such as the secondary metabolites, cell motility and antibiotic resistance are all influenced by QS [58][59]; in addition, QS signals that contribute to the interactions between planktonic microalgae and bacteria are summarized as the N-acyl-homoserine lactones (AHLs) [61], the 2-alkyl-4-quinolones (AQs) [59], long-chain fatty acids and fatty acid methyl esters (autoinducer-2, AI-2) and dihydroxypentanedione furanone derivates [12]. It is agreed that most of the anticyanobacterial activities by Gram-negative bacteria (such as Pseudomonas sp., Acinetobacter sp., etc.) are the consequence of bacterial-cyanobacterial QS rather than bacterium-cyanobacteria interactions [12][60]. Some species of Serratia sp. [63] and Hahella sp. [48] can produce prodigiosin to inhibit M. aeruginosa, and the prodigiosin production is regulated by LuxI and LuxR, which are the crucial genes of AHLs [64]. The QS signal molecule (C4-HSL), which belongs to the classic AHL-based LuxIR-type QS system of Gram-negative bacteria, is responsible for the synthetic process of the anticyanobacterial compound (3-methylindole) from Aeromonas sp. GLY-2107 [61]. During the anticyanobacterial process, the QS systems of Gram-negative bacteria produce AHLs signaling molecules, which are synthesized by the basic regulatory protein of LuxI [61][64][65].
In contrast, a wide range of the Gram-positive anticyanobacteria (such as Streptomyces sp., Bacillus sp., etc.) generally use AI-2 as the signal molecules in QS systems [62]. The anticyanobacterium S. xiamenensis Lzh-2 exhibits QS behavior, and the LuxS gene is crucial for the AI-2 type QS system; obviously, the anticyanobacterial activity of S. xiamenensis Lzh-2 is regulated through the LuxS/AI-2 QS system by inducing the production of anticyanobacterial compounds 2, 3-indolinedione and cyclo(Gly-Pro) [64]. The AI-2 type QS behavior is present in Bacillus sp. [66]. Genomic analysis of B. subtilis JA has indicated the existence of the LuxS gene that regulates the pheromone biosynthesis, and the high-molecular-weight anticyanobacterial compounds (>3 kDa) produced by Bacillus sp. S51107 have been proven to be primarily regulated by the NprR-NprX-type (AI-2) QS system [65]. As a consequence, the AI-2 QS system has been considered as a possible strategy to regulate the behavior of the anticyanobacterial effects of Gram-positive bacteria. Although QS behavior has been reported in recent years, there is still an improved understanding of the interaction between cyanobacteria and anticyanobacterial microorganisms.
References
- Harke, M.J.; Steffen, M.M.; Gobler, C.J.; Otten, T.G.; Wilhelm, S.; Wood, S.A.; Paerl, H.W. A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 2016, 54, 4–20.
- Yang, C.; Hou, X.; Wu, D.; Chang, W.; Zhang, X.; Dai, X.; Du, H.; Zhang, X.; Igarashi, Y.; Luo, F. The characteristics and algicidal mechanisms of cyanobactericidal bacteria, a review. World J. Microbiol. Biotechnol. 2020, 36, 188.
- Ko, S.-R.; Lee, Y.-K.; Srivastava, A.; Park, S.-H.; Ahn, C.-Y.; Oh, H.-M. The Selective Inhibitory Activity of a Fusaricidin Derivative on a Bloom-Forming Cyanobacterium, Microcystis sp. J. Microbiol. Biotechnol. 2019, 29, 59–65.
- Han, S.-I.; Kim, S.; Choi, K.Y.; Lee, C.; Park, Y.; Choi, Y.-E. Control of a toxic cyanobacterial bloom species, Microcystis aeruginosa, using the peptide HPA3NT3-A2. Environ. Sci. Pollut. Res. 2019, 26, 32255–32265.
- Yu, Y.; Zeng, Y.; Li, J.; Yang, C.; Zhang, X.; Luo, F.; Dai, X. An algicidal Streptomyces amritsarensis strain against Microcystis aeruginosa strongly inhibits microcystin synthesis simultaneously. Sci. Total Environ. 2018, 650, 34–43.
- Mohamed, Z.A.; Hashem, M.; Alamri, S.A. Growth inhibition of the cyanobacterium Microcystis aeruginosa and degradation of its microcystin toxins by the fungus Trichoderma citrinoviride. Toxicon 2014, 86, 51–58.
- Goslan, E.H.; Seigle, C.; Purcell, D.; Henderson, R.; Parsons, S.A.; Jefferson, B.; Judd, S.J. Carbonaceous and nitrogenous disinfection by-product formation from algal organic matter. Chemosphere 2016, 170, 1–9.
- Xin, H.; Yang, S.; Tang, Y.; Wu, M.; Deng, Y.; Xu, B.; Gao, N. Mechanisms and performance of calcium peroxide-enhanced Fe(ii) coagulation for treatment of Microcystis aeruginosa-laden water. Environ. Sci. Water Res. Technol. 2020, 6, 1272–1285.
- Chen, Z.; Li, J.; Chen, M.; Koh, K.Y.; Du, Z.; Gin, K.Y.-H.; He, Y.; Ong, C.N.; Chen, J.P. Microcystis aeruginosa removal by peroxides of hydrogen peroxide, peroxymonosulfate and peroxydisulfate without additional activators. Water Res. 2021, 201, 117263.
- Wang, M.; Chen, S.; Zhou, W.; Yuan, W.; Wang, D. Algal cell lysis by bacteria: A review and comparison to conventional methods. Algal Res. 2020, 46, 101794.
- Matthijs, H.C.P.; Jančula, D.; Visser, P.M.; Maršálek, B. Existing and emerging cyanocidal compounds: New perspectives for cyanobacterial bloom mitigation. Aquat. Ecol. 2016, 50, 443–460.
- Demuez, M.; González-Fernández, C.; Ballesteros, M. Algicidal microorganisms and secreted algicides: New tools to induce microalgal cell disruption. Biotechnol. Adv. 2015, 33, 1615–1625.
- Sun, R.; Sun, P.; Zhang, J.; Esquivel-Elizondo, S.; Wu, Y. Microorganisms-based methods for harmful algal blooms control: A review. Bioresour. Technol. 2018, 248, 12–20.
- Benegas, G.R.S.; Bernal, S.P.F.; de Oliveira, V.M.; Passarini, M.R.Z. Antimicrobial activity against Microcystis aeruginosa and degradation of microcystin-LR by bacteria isolated from Antarctica. Environ. Sci. Pollut. Res. 2021, 28, 52381–52391.
- Li, Y.; Wu, X.; Jiang, X.; Liu, L.; Wang, H. Algicidal activity of Aspergillus niger induced by calcium ion as signal molecule on Microcystis aeruginosa. Algal Res. 2021, 60, 102536.
- Kong, Y.; Wang, Q.; Chen, Y.; Xu, X.; Zhu, L.; Yao, H.; Pan, H. Anticyanobacterial process and action mechanism of Streptomyces sp. HJC-D1 on Microcystis aeruginosa. Environ. Prog. Sustain. Energy 2020, 39, e13392.
- Yi, Y.-L.; Yu, X.-B.; Zhang, C.; Wang, G.-X. Growth inhibition and microcystin degradation effects of Acinetobacter guillouiae A2 on Microcystis aeruginosa. Res. Microbiol. 2015, 166, 93–101.
- Gerphagnon, M.; Macarthur, D.; Latour, D.; Gachon, C.; Van Ogtrop, F.; Gleason, F.H.; Sime-Ngando, T. Microbial players involved in the decline of filamentous and colonial cyanobacterial blooms with a focus on fungal parasitism. Environ. Microbiol. 2015, 17, 2573–2587.
- Shunyu, S.; Yongding, L.; Yinwu, S.; Genbao, L.; Dunhai, L. Lysis of Aphanizomenon flos-aquae (Cyanobacterium) by a bacterium Bacillus cereus. Biol. Control 2006, 39, 345–351.
- Park, B.S.; Park, C.-S.; Shin, Y.; Yoon, S.; Han, M.-S.; Kang, Y.-H. Different Algicidal Modes of the Two Bacteria Aeromonas bestiarum HYD0802-MK36 and Pseudomonas syringae KACC10292T against Harmful Cyanobacteria Microcystis aeruginosa. Toxins 2022, 14, 128.
- Zhang, C.; Massey, I.Y.; Liu, Y.; Huang, F.; Gao, R.; Ding, M.; Xiang, L.; He, C.; Wei, J.; Li, Y.; et al. Identification and characterization of a novel indigenous algicidal bacterium Chryseobacterium species against Microcystis aeruginosa. J. Toxicol. Environ. Heal. Part A 2019, 82, 845–853.
- Zeng, Y.; Wang, J.; Yang, C.; Ding, M.; Hamilton, P.B.; Zhang, X.; Yang, C.; Zhnag, L.; Dai, X. A Streptomyces globisporus strain kills Microcystis aeruginosa via cell-to-cell contact. Sci. Total Environ. 2021, 769, 144489.
- Pathmalal, M.M.; Zenichiro, K.; Shin-ichi, N. Algicidal effect of the bacterium Alcaligenes denitrificans on Microcystis spp. Aquatic Microbial Ecology 2000, 22, 111–117.
- Xue, G.; Wang, X.; Xu, C.; Song, B.; Chen, H. Removal of harmful algae by Shigella sp. H3 and Alcaligenes sp. H5: Algicidal pathways and characteristics. Environ. Technol. 2021.
- Li, H.; Ai, H.; Kang, L.; Sun, X.; He, Q. Simultaneous Microcystis Algicidal and Microcystin Degrading Capability by a Single Acinetobacter Bacterial Strain. Environ. Sci. Technol. 2016, 50, 11903–11911.
- Su, J.F.; Shao, S.C.; Huang, T.L.; Ma, F.; Lu, J.S.; Zhang, K. Algicidal effects and denitrification activities of Acinetobacter sp. J25 against Microcystis aeruginosa. J. Environ. Chem. Eng. 2016, 4, 1002–1007.
- Li, Y.; Liu, L.; Xu, Y.; Li, P.; Zhang, K.; Jiang, X.; Zheng, T.; Wang, H. Stress of algicidal substances from a bacterium Exiguobacterium sp. h10 on Microcystis aeruginosa. Lett. Appl. Microbiol. 2016, 64, 57–65.
- Zhang, S.; Fan, C.; Xia, Y.; Li, M.; Wang, Y.; Cui, X.; Xiao, W. Characterization of a novel bacteriophage specific to Exiguobacterium indicum isolated from a plateau eutrophic lake. J. Basic Microbiol. 2018, 59, 206–214.
- Tian, C.; Liu, X.; Tan, J.; Lin, S.; Li, D.; Yang, H. Isolation, identification and characterization of an algicidal bacterium from Lake Taihu and preliminary studies on its algicidal compounds. J. Environ. Sci. 2012, 24, 1823–1831.
- Wu, L.; Wu, H.; Chen, L.; Xie, S.; Zang, H.; Borriss, R.; Gao, X. Bacilysin from Bacillus amyloliquefaciens FZB42 Has Specific Bactericidal Activity against Harmful Algal Bloom Species. Appl. Environ. Microbiol. 2014, 80, 7512–7520.
- Yu, J.; Kong, Y.; Gao, S.; Miao, L.; Zou, P.; Xu, B.; Zeng, C.; Zhang, X. Bacillus amyloliquefaciens T1 as a potential control agent for cyanobacteria. J. Appl. Phycol. 2014, 27, 1213–1221.
- Xu, B.; Miao, L.; Yu, J.; Ji, L.; Lu, H.; Yang, J.; Gao, S.; Kong, Y. Isolation and identification of amino acids secreted by Bacillus amyloliquefaciens T1 with anti-cyanobacterial effect against cyanobacterium Microcystis aeruginosa. Desalination Water Treat. 2021, 231, 329–339.
- Chen, W.-M.; Sheu, F.-S.; Sheu, S.-Y. Aquimarina salinaria sp. nov., a novel algicidal bacterium isolated from a saltpan. Arch. Microbiol. 2011, 194, 103–112.
- Van Le, V.; Ko, S.-R.; Kang, M.; Lee, S.-A.; Oh, H.-M.; Ahn, C.-Y. Algicide capacity of Paucibacter aquatile DH15 on Microcystis aeruginosa by attachment and non-attachment effects. Environ. Pollut. 2022, 302, 119079.
- Chen, Y.-D.; Zhu, Y.; Xin, J.-P.; Zhao, C.; Tian, R.-N. Succinic acid inhibits photosynthesis of Microcystis aeruginosa via damaging PSII oxygen-evolving complex and reaction center. Environ. Sci. Pollut. Res. 2021, 28, 58470–58479.
- Kong, Y.; Zou, P.; Yang, Q.; Xu, X.; Miao, L.; Zhu, L. Physiological responses of Microcystis aeruginosa under the stress of antialgal actinomycetes. J. Hazard. Mater. 2013, 262, 274–280.
- Li, D.; Kang, X.; Chu, L.; Wang, Y.; Song, X.; Zhao, X.; Cao, X. Algicidal mechanism of Raoultella ornithinolytica against Microcystis aeruginosa: Antioxidant response, photosynthetic system damage and microcystin degradation. Environ. Pollut. 2021, 287, 117644.
- Shao, J.; Jiang, Y.; Wang, Z.; Peng, L.; Luo, S.; Gu, J.; Li, R. Interactions between algicidal bacteria and the cyanobacterium Microcystis aeruginosa: Lytic characteristics and physiological responses in the cyanobacteria. Int. J. Environ. Sci. Technol. 2013, 11, 469–476.
- Zhou, S.; Yin, H.; Tang, S.; Peng, H.; Yin, D.; Yang, Y.; Liu, Z.; Dang, Z. Physiological responses of Microcystis aeruginosa against the algicidal bacterium Pseudomonas aeruginosa. Ecotoxicol. Environ. Saf. 2016, 127, 214–221.
- Wang, X.; Xie, M.; Wu, W.; Shi, L.; Luo, L.; Li, P. Differential sensitivity of colonial and unicellular Microcystis strains to an algicidal bacterium Pseudomonas aeruginosa. J. Plankton Res. 2013, 35, 1172–1176.
- Luo, J.; Wang, Y.; Tang, S.; Liang, J.; Lin, W.; Luo, L. Isolation and Identification of Algicidal Compound from Streptomyces and Algicidal Mechanism to Microcystis aeruginosa. PLoS ONE 2013, 8, e76444.
- Hua, X.-H.; Li, J.-H.; Li, J.-J.; Zhang, L.-H.; Cui, Y. Selective inhibition of the cyanobacterium, Microcystis, by a Streptomyces sp. Biotechnol. Lett. 2009, 31, 1531–1535.
- Guo, X.; Liu, X.; Pan, J.; Yang, H. Synergistic algicidal effect and mechanism of two diketopiperazines produced by Chryseobacterium sp. strain GLY-1106 on the harmful bloom-forming Microcystis aeruginosa. Sci. Rep. 2015, 5, 14720.
- Wang, S.; Yang, S.; Zuo, J.; Hu, C.; Song, L.; Gan, N.; Chen, P. Simultaneous Removal of the Freshwater Bloom-Forming Cyanobacterium Microcystis and Cyanotoxin Microcystins via Combined Use of Algicidal Bacterial Filtrate and the Microcystin-Degrading Enzymatic Agent, MlrA. Microorganisms 2021, 9, 1594.
- Liu, H.; Guo, X.; Liu, L.; Yan, M.; Li, J.; Hou, S.; Wan, J.; Feng, L. Simultaneous Microcystin Degradation and Microcystis aeruginosa Inhibition with the Single Enzyme Microcystinase A. Environ. Sci. Technol. 2020, 54, 8811–8820.
- Xuan, H.; Dai, X.; Li, J.; Zhang, X.; Yang, C.; Luo, F. A Bacillus sp. strain with antagonistic activity against Fusarium graminearum kills Microcystis aeruginosa selectively. Sci. Total Environ. 2017, 583, 214–221.
- Kong, Y.; Xu, X.; Zhu, L. Cyanobactericidal Effect of Streptomyces sp. HJC-D1 on Microcystis auruginosa. PLoS ONE 2013, 8, e57654.
- Yang, K.; Chen, Q.; Zhang, D.; Zhang, H.; Lei, X.; Chen, Z.; Li, Y.; Hong, Y.; Ma, X.; Zheng, W.; et al. The algicidal mechanism of prodigiosin from Hahella sp. KA22 against Microcystis aeruginosa. Sci. Rep. 2017, 7, 7750.
- Zhang, B.-H.; Ding, Z.-G.; Li, H.-Q.; Mou, X.-Z.; Zhang, Y.-Q.; Yang, J.-Y.; Zhou, E.-M.; Li, W.-J. Algicidal Activity of Streptomyces eurocidicus JXJ-0089 Metabolites and Their Effects on Microcystis Physiology. Appl. Environ. Microbiol. 2016, 82, 5132–5143.
- Jin, P.; Wang, H.; Liu, W.; Zhang, S.; Lin, C.; Zheng, F.; Miao, W. Bactericidal metabolites from Phellinus noxius HN-1 against Microcystis aeruginosa. Sci. Rep. 2017, 7, 3132.
- Zeng, G.; Zhang, M.; Gao, P.; Wang, J.; Sun, D. Algicidal Efficiency and Genotoxic Effects of Phanerochaete chrysosporium against Microcystis aeruginosa. Int. J. Environ. Res. Public Health 2020, 17, 4029.
- Zhang, X.; Song, T.; Ma, H.; Li, L. Physiological response of Microcystis aeruginosa to the extracellular substances from an Aeromonas sp. RSC Adv. 2016, 6, 103662–103667.
- Liu, J.; Yang, C.; Chi, Y.; Wu, D.; Dai, X.; Zhang, X.; Igarashi, Y.; Luo, F. Algicidal characterization and mechanism of Bacillus licheniformis Sp34 against Microcystis aeruginosa in Dianchi Lake. J. Basic Microbiol. 2019, 59, 1112–1124.
- Chen, Q.; Wang, L.; Qi, Y.; Ma, C. Imaging mass spectrometry of interspecies metabolic exchange revealed the allelopathic interaction between Microcystis aeruginosa and its antagonist. Chemosphere 2020, 259, 127430.
- Zhang, Y.; Chen, D.; Zhang, N.; Li, F.; Luo, X.; Li, Q.; Li, C.; Huang, X. Transcriptional Analysis of Microcystis aeruginosa Co-Cultured with Algicidal Bacteria Brevibacillus laterosporus. Int. J. Environ. Res. Public Health 2021, 18, 8615.
- Zeng, G.; Gao, P.; Wang, J.; Zhang, J.; Zhang, M.; Sun, D. Algicidal Molecular Mechanism and Toxicological Degradation of Microcystis aeruginosa by White-Rot Fungi. Toxins 2020, 12, 406.
- Han, S.; Zhou, Q.; Lilje, O.; Xu, W.; Zhu, Y.; van Ogtrop, F.F. Inhibition mechanism of Penicillium chrysogenum on Microcystis aeruginosa in aquaculture water. J. Clean. Prod. 2021, 299, 126829.
- Zhai, C.; Zhang, P.; Shen, F.; Zhou, C.; Liu, C. Does Microcystis aeruginosa have quorum sensing? FEMS Microbiol. Lett. 2012, 336, 38–44.
- Reading, N.C.; Sperandio, V. Quorum sensing: The many languages of bacteria. FEMS Microbiol. Lett. 2006, 254, 1–11.
- Zhang, Y.; Zheng, L.; Wang, S.; Zhao, Y.; Xu, X.; Han, B.; Hu, T. Quorum Sensing Bacteria in the Phycosphere of HAB Microalgae and Their Ecological Functions Related to Cross-Kingdom Interactions. Int. J. Environ. Res. Public Health 2021, 19, 163.
- Guo, X.; Liu, X.; Wu, L.; Pan, J.; Yang, H. The algicidal activity of Aeromonas sp. strain GLY-2107 against bloom-forming Microcystis aeruginosa is regulated by N-acyl homoserine lactone-mediated quorum sensing. Environ. Microbiol. 2016, 18, 3867–3883.
- Dow, L. How Do Quorum-Sensing Signals Mediate Algae–Bacteria Interactions? Microorganisms 2021, 9, 1391.
- Wei, J.; Xie, X.; Huang, F.; Xiang, L.; Wang, Y.; Han, T.; Massey, I.Y.; Liang, G.; Pu, Y.; Yang, F. Simultaneous Microcystis algicidal and microcystin synthesis inhibition by a red pigment prodigiosin. Environ. Pollut. 2019, 256, 113444.
- Liu, J.; Liu, K.; Zhao, Z.; Wang, Z.; Wang, F.; Xin, Y.; Qu, J.; Song, F.; Li, Z. The LuxS/AI-2 Quorum-Sensing System Regulates the Algicidal Activity of Shewanella xiamenensis Lzh-2. Front. Microbiol. 2022, 12.
- Wu, L.; Guo, X.; Liu, X.; Yang, H. NprR-NprX Quorum-Sensing System Regulates the Algicidal Activity of Bacillus sp. Strain S51107 against Bloom-Forming Cyanobacterium Microcystis aeruginosa. Front. Microbiol. 2017, 8, 1968.
- Zhang, S.-J.; Du, X.-P.; Zhu, J.-M.; Meng, C.-X.; Zhou, J.; Zuo, P. The complete genome sequence of the algicidal bacterium Bacillus subtilis strain JA and the use of quorum sensing to evaluate its antialgal ability. Biotechnol. Rep. 2020, 25, e00421.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
906
Revisions:
2 times
(View History)
Update Date:
20 Jun 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




