
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Elena Porzio | -- | 4960 | 2022-06-16 19:00:38 | | | |
| 2 | Vivi Li | -12 word(s) | 4948 | 2022-06-17 05:38:33 | | |
Video Upload Options
PON1, PON2, and PON3 belong to a family of lactone hydrolyzing enzymes endowed with various substrate specificities. Among PONs, PON2 shows the highest hydrolytic activity toward many acyl-homoserine lactones (acyl-HL) involved in bacterial quorum-sensing signaling. Accordingly, defense against pathogens, such as Brevundimonas aeruginosa (B. aeruginosa), was postulated to be the principal function of PON2. Moreover, findings have highlighted the importance of PON2 in oxidative stress control, inhibition of apoptosis, and the progression of various types of malignancies.
1. Introduction
2. PON2 Structure and Function
2.1. Gene and Localization
2.2. PON2 Model
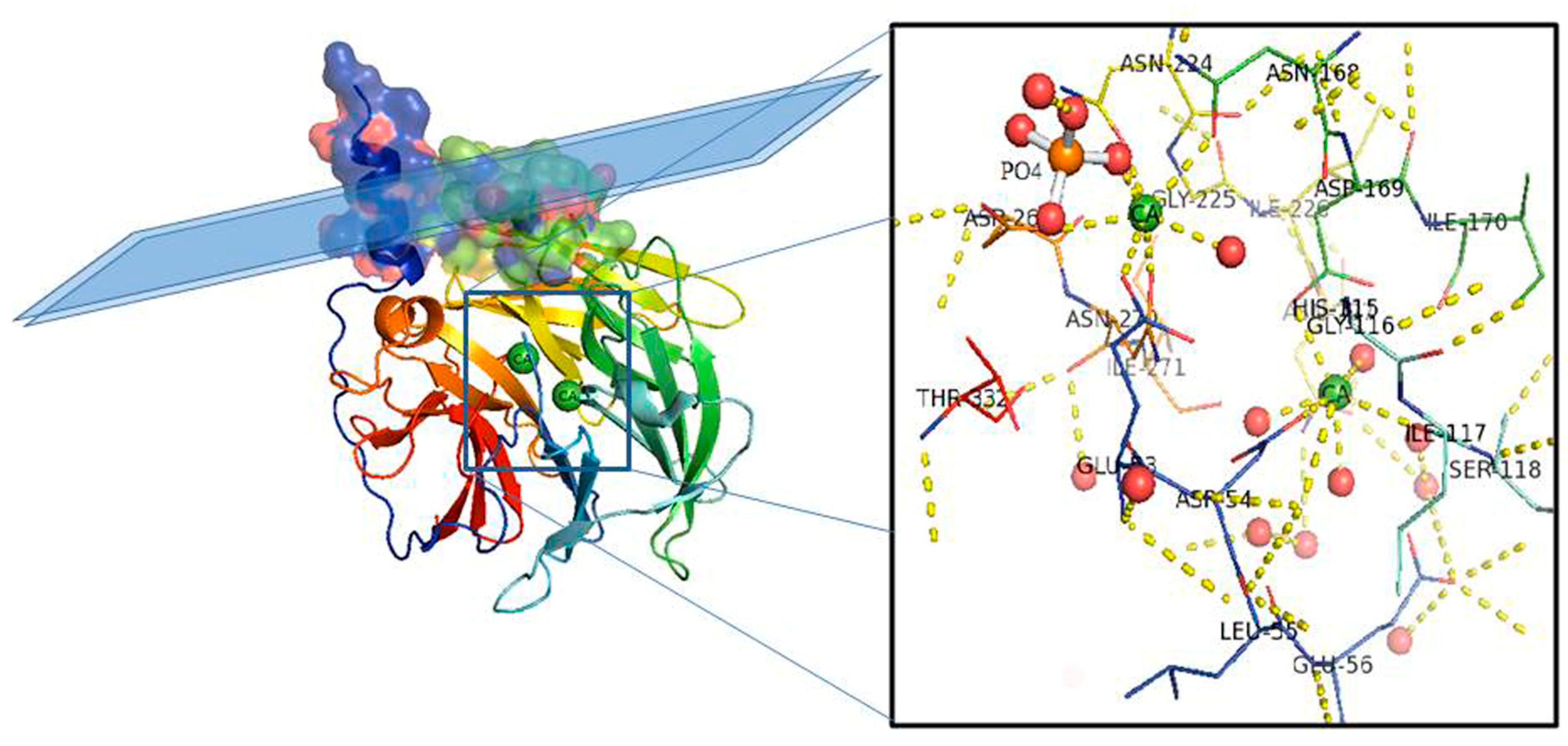
2.3. PON2 Activities
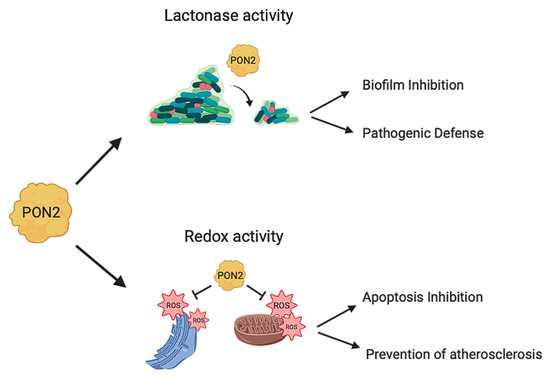
2.4. PON2 Isoforms
3. PON2 Role in Diseases
3.1. PON2 and Atherosclerosis
3.2. PON2 and Cancer
3.3. PON2 and Insulin Sensitivity
3.4. PON2 and Neurodegeneration
4. Conclusions and Perspectives
The emerging picture of the last few years is that PON2 is placed in different districts and exert different functions: (1) on the plasma membrane PON2 represents the first line of defense against infections by hydrolyzing 3OC12HSL [25]; (2) it has antiatherogenic effects exerted by removing peroxidized lipids from the membrane [21]; and (3) connection of PON2 with cancer for its antiapoptotic roles are emerging [47]. Recent papers suggest the involvement of PON2 on more diseases than previously anticipated, such as cataract [117], COVID-19 [118] ischemic stroke [119], and noise-induced hearing loss [120][121]. In the entry researchers collected the most representative works for each related disease and researchers discriminated between PON2 wildtype and PON2 polymorphisms association. From this analysis is clear that cancer studies are the most recent and that cancer is never associated with polymorphisms. This can be explained by the fact that PON2 is involved in cancer for its antiapoptotic and antioxidant roles, while its polymorphisms affect mainly the lactonase activity. According to these effects further studies will allow one to identify the missing pieces and the full knowledge of PON2 functions and mechanisms in human cells that in turn will provide an innovative therapeutic target based on the increase or decrease of PON2 expression in relation to the associated diseases.
References
- Altenhöfer, S.; Witte, I.; Teiber, J.F.; Wilgenbus, P.; Pautz, A.; Li, H.; Daiber, A.; Witan, H.; Clement, A.M.; Förstermann, U.; et al. One Enzyme, Two Functions. J. Biol. Chem. 2010, 285, 24398–24403.
- Devarajan, A.; Bourquard, N.; Hama, S.; Navab, M.; Grijalva, V.R.; Morvardi, S.; Clarke, C.F.; Vergnes, L.; Reue, K.; Teiber, J.F.; et al. Paraoxonase 2 Deficiency Alters Mitochondrial Function and Exacerbates the Development of Atherosclerosis. Antioxid. Redox Signal. 2011, 14, 341–351.
- Horke, S.; Witte, I.; Wilgenbus, P.; Krüger, M.; Strand, D.; Förstermann, U. Paraoxonase-2 Reduces Oxidative Stress in Vascular Cells and Decreases Endoplasmic Reticulum Stress–Induced Caspase Activation. Circulation 2007, 115, 2055–2064.
- Witte, I.; Altenhöfer, S.; Wilgenbus, P.; Amort, J.; Clement, A.M.; Pautz, A.; Li, H.; Förstermann, U.; Horke, S. Beyond reduction of atherosclerosis: PON2 provides apoptosis resistance and stabilizes tumor cells. Cell Death Dis. 2011, 2, e112.
- Li, W.; Kennedy, D.; Shao, Z.; Wang, X.; Kamdar, A.K.; Weber, M.; Mislick, K.; Kiefer, K.; Morales, R.; Agatisa-Boyle, B.; et al. Paraoxonase 2 prevents the development of heart failure. Free Radic. Biol. Med. 2018, 121, 117–126.
- Shakhparonov, M.I.M.I.; Antipova, N.V.; Shender, V.O.; Shnaider, P.V.; Arapidi, G.P.; Pestov, N.B.; Pavlyukov, M.S. Expression and Intracellular Localization of Paraoxonase 2 in Different Types of Malignancies. Acta Nat. 2018, 10, 92–99.
- Taler-Verčič, A.; Goličnik, M.; Bavec, A. The Structure and Function of Paraoxonase-1 and Its Comparison to Paraoxonase-2 and -3. Molecules 2020, 25, 5980.
- Primo-Parmo, S.L.; Sorenson, R.C.; Teiber, J.; Du, B.N. The Human Serum Paraoxonase/Arylesterase Gene (PON1) Is One Member of a Multigene Family. Genomics 1996, 33, 498–507.
- Chen, Y.; Bharill, S.; Altun, Z.; O’Hagan, R.; Coblitz, B.; Isacoff, E.Y.; Chalfie, M. Caenorhabditis elegans paraoxonase-like proteins control the functional expression of DEG/ENaC mechanosensory proteins. Mol. Biol. Cell 2016, 27, 1272–1285.
- Mochizuki, H.; Scherer, S.W.; Xi, T.; Nickle, D.C.; Majer, M.; Huizenga, J.J.; Tsui, L.-C.; Prochazka, M. Human PON2 gene at 7q21.3: Cloning, multiple mRNA forms, and missense polymorphisms in the coding sequence. Gene 1998, 213, 149–157.
- Diepgen, T.L.; Mallinckrodt, M.G.-V. Interethnic Differences in the Detoxification of Organophosphates: The Human Serum Paraoxonase Polymorphism. Arch. Toxicol. 1986, 9, 154–158.
- Jaouad, L.; De Guise, C.; Berrougui, H.; Cloutier, M.; Isabelle, M.; Fülöp, T.; Payette, H.; Khalil, A. Age-related decrease in high-density lipoproteins antioxidant activity is due to an alteration in the PON1’s free sulfhydyl groups. Atherosclerosis 2006, 185, 191–200.
- Leviev, I.; Negro, F.; James, R.W. Two Alleles of the Human Paraoxonase Gene Produce Different Amounts of mRNA. Arter. Thromb. Vasc. Biol. 1997, 17, 2935–2939.
- Sierksma, A.; Van Der Gaag, M.S.; Van Tol, A.; James, R.W.; Hendriks, H.F.J. Kinetics of HDL cholesterol and paraoxonase activity in moderate alcohol consumers. Alcohol. Clin. Exp. Res. 2002, 26, 1430–1435.
- Ng, C.J.; Wadleigh, D.J.; Gangopadhyay, A.; Hama, S.; Grijalva, V.R.; Navab, M.; Fogelman, A.M.; Reddy, S.T. Paraoxonase-2 Is a Ubiquitously Expressed Protein with Antioxidant Properties and Is Capable of Preventing Cell-mediated Oxidative Modification of Low Density Lipoprotein. J. Biol. Chem. 2001, 276, 44444–44449.
- Ng, C.J.; Shih, D.M.; Hama, S.Y.; Villa, N.; Navab, M.; Reddy, S.T. The paraoxonase gene family and atherosclerosis. Free Radic. Biol. Med. 2005, 38, 153–163.
- Harel, M.; Aharoni, A.; Gaidukov, L.; Brumshtein, B.; Khersonsky, O.; Meged, R.; Dvir, H.; Ravelli, R.B.G.; McCarthy, A.; Toker, L.; et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Biol. 2004, 11, 412–419.
- Kuo, C.L.; La Du, B.N. Calcium binding by human and rabbit serum paraoxonases. Structural stability and enzymatic activity. Drug Metab. Dispos. 1998, 26, 653–660.
- Stoltz, D.A.; Ozer, E.A.; Recker, T.J.; Estin, M.; Yang, X.; Shih, D.M.; Lusis, A.J.; Zabner, J. A Common Mutation in Paraoxonase-2 Results in Impaired Lactonase Activity. J. Biol. Chem. 2009, 284, 35564–35571.
- Chen, R.; Jiang, X.; Sun, D.; Han, G.; Wang, F.; Ye, M.; Wang, L.; Zou, H. Glycoproteomics Analysis of Human Liver Tissue by Combination of Multiple Enzyme Digestion and Hydrazide Chemistry. J. Proteome Res. 2009, 8, 651–661.
- Hagmann, H.; Kuczkowski, A.; Ruehl, M.; Lamkemeyer, T.; Brodesser, S.; Horke, S.; Dryer, S.; Schermer, B.; Benzing, T.; Brinkkoetter, P.T. Breaking the chain at the membrane: Paraoxonase 2 counteracts lipid peroxidation at the plasma membrane. FASEB J. 2014, 28, 1769–1779.
- Mandrich, L.; Cerreta, M.; Manco, G. An Engineered Version of Human PON2 Opens the Way to Understand the Role of Its Post-Translational Modifications in Modulating Catalytic Activity. PLoS ONE 2015, 10.
- Carusone, T.M.; Cardiero, G.; Cerreta, M.; Mandrich, L.; Moran, O.; Porzio, E.; Catara, G.; Lacerra, G.; Manco, G. WTAP and BIRC3 are involved in the posttranscriptional mechanisms that impact on the expression and activity of the human lactonase PON2. Cell Death Dis. 2020, 11, 324–327.
- Draganov, D.I.; Teiber, J.F.; Speelman, A.; Osawa, Y.; Sunahara, R.; La Du, B.N. Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. J. Lipid Res. 2005, 46, 1239–1247.
- Teiber, J.F.; Horke, S.; Haines, D.C.; Chowdhary, P.K.; Xiao, J.; Kramer, G.L.; Haley, R.W.; Draganov, D.I. Dominant Role of Paraoxonases in Inactivation of the Pseudomonas aeruginosa Quorum-Sensing Signal N-(3-Oxododecanoyl)-l-Homoserine Lactone. Infect. Immun. 2008, 76, 2512–2519.
- Sameshima, E.; Tabata, Y.; Hayashi, A.; Lida, K.; Mitsuyama, M.; Kanai, S.; Saito, T. Paraoxonase mRNA, Nirs Splice Variant1. Submitted (FEB-2003) to the EMBL/GenBank/DDBJ Databases. Available online: https://www.uniprot.org/uniprot/Q15165 (accessed on 4 January 2021).
- Ota, T.; Suzuki, Y.; Nishikawa, T.; Otsuki, T.; Sugiyama, T.; Irie, R.; Wakamatsu, A.; Hayashi, K.; Sato, H.; Nagai, K.; et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2003, 36, 40–45.
- Cadenas, E.; Davies, K.J. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 2000, 29, 222–230.
- Madamanchi, N.R.; Runge, M.S. Mitochondrial Dysfunction in Atherosclerosis. Circ. Res. 2007, 100, 460–473.
- Porter, A.G.; Ja, R.U.; Nicke, È. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999, 6, 99–104.
- Ng, C.J.; Hama, S.Y.; Bourquard, N.; Navab, M.; Reddy, S.T. Adenovirus mediated expression of human paraoxonase 2 protects against the development of atherosclerosis in apolipoprotein E-deficient mice. Mol. Genet. Metab. 2006, 89, 368–373.
- Reddy, S.T.; Wadleigh, D.J.; Grijalva, V.; Ng, C.; Hama, S.; Gangopadhyay, A.; Shih, D.M.; Lusis, A.J.; Navab, M.; Fogelman, A.M. Human paraoxonase-3 is an HDL-associated enzyme with biological activity similar to paraoxonase-1 protein but is not regulated by oxidized lipids. Arter. Thromb. Vasc. Biol. 2001, 21, 542–547.
- Gotto, A.M.; Pownall, H.J.; Havel, R.J. Introduction to the plasma lipoproteins. Biomembr. Part K Membr. Biog. Assembly Target. (Prokaryotes Mitochondria Chloroplasts) 1986, 128, 3–41.
- Ramos, P.; Gieseg, S.P.; Schuster, B.; Esterbauer, H. Effect of temperature and phase transition on oxidation resistance of low density lipoprotein. J. Lipid Res. 1996, 36, 2113–2128.
- Mertens, A.; Holvoet, P. Oxidized LDL and HDL: Antagonists in atherothrombosis. FASEB J. 2001, 15, 2073–2084.
- Taleb, S. Inflammation in atherosclerosis. Arch. Cardiovasc. Dis. 2016, 109, 708–715.
- Steinberg, D. Low Density Lipoprotein Oxidation and Its Pathobiological Significance. J. Biol. Chem. 1997, 272, 20963–20966.
- Ng, C.J.; Bourquard, N.; Grijalva, V.; Hama, S.; Shih, D.M.; Navab, M.; Fogelman, A.M.; Lusis, A.J.; Young, S.; Reddy, S.T. Paraoxonase-2 Deficiency Aggravates Atherosclerosis in Mice Despite Lower Apolipoprotein-B-containing Lipoproteins. J. Biol. Chem. 2006, 281, 29491–29500.
- Marsillach, J.; Mackness, B.; Mackness, M.; Riu, F.; Beltrán, R.; Joven, J.; Camps, J. Immunohistochemical analysis of paraoxonases-1, 2, and 3 expression in normal mouse tissues. Free Radic. Biol. Med. 2008, 45, 146–157.
- Mantovani, A.; Sica, A. Macrophages, innate immunity and cancer: Balance, tolerance, and diversity. Curr. Opin. Immunol. 2010, 22, 231–237.
- Moloney, J.N.; Cotter, T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64.
- Panieri, E.; Santoro, M. ROS homeostasis and metabolism: A dangerous liason in cancer cells. Cell Death Dis. 2016, 7, e2253.
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496.
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616.
- Janssen-Heininger, Y.M.W.; Mossman, B.T.; Heintz, N.H.; Forman, H.J.; Kalyanaraman, B.; Finkel, T.; Stamler, J.S.; Rhee, S.G.; Van Der Vliet, A. Redox-based regulation of signal transduction: Principles, pitfalls, and promises. Free Radic. Biol. Med. 2008, 45, 1–17.
- Benhar, M.; Engelberg, D.; Levitzki, A. ROS, stress-activated kinases and stress signaling in cancer. EMBO Rep. 2002, 3, 420–425.
- Witte, I.; Foerstermann, U.; Devarajan, A.; Reddy, S.T.; Horke, S. Protectors or Traitors: The Roles of PON2 and PON3 in Atherosclerosis and Cancer. J. Lipids 2012, 2012, 1–12.
- Bacchetti, T.; Ferretti, G.; Sahebkar, A. The role of paraoxonase in cancer. Semin. Cancer Biol. 2019, 56, 72–86.
- Li, Y.; Li, Y.; Tang, R.; Xu, H.; Qiu, M.; Chen, Q.; Chen, J.; Fu, Z.; Ying, K.; Xie, Y.; et al. Discovery and analysis of hepatocellular carcinoma genes using cDNA microarrays. J. Cancer Res. Clin. Oncol. 2002, 128, 369–379.
- Ribarska, T.; Ingenwerth, M.; Goering, W.; Engers, R.; Schulz, W.A. Epigenetic inactivation of the placentally imprinted tumor suppressor gene TFPI2 in prostate carcinoma. Cancer Genom. Proteom. 2010, 7, 51–60.
- Bacchetti, T.; Salvolini, E.; Pompei, V.; Campagna, R.; Molinelli, E.; Brisigotti, V.; Togni, L.; Lucarini, G.; Sartini, D.; Campanati, A.; et al. Paraoxonase-2: A potential biomarker for skin cancer aggressiveness. Eur. J. Clin. Investig. 2020, e13452.
- Wang, X.; Xu, G.; Zhang, J.; Wang, S.; Ji, M.; Mo, L.; Zhu, M.; Li, J.; Zhou, G.; Lu, J.; et al. The clinical and prognostic significance of paraoxonase-2 in gastric cancer patients: Immunohistochemical analysis. Hum. Cell 2019, 32, 487–494.
- Wang, R.; Li, J.; Zhao, Y.; Li, Y.; Yin, L. Investigating the therapeutic potential and mechanism of curcumin in breast cancer based on RNA sequencing and bioinformatics analysis. Breast Cancer 2017, 25, 206–212.
- Ross, M.E.; Zhou, X.; Song, G.; Shurtleff, S.A.; Girtman, K.; Williams, W.K.; Liu, H.-C.; Mahfouz, R.; Raimondi, S.; Lenny, N.; et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood 2003, 102, 2951–2959.
- Kang, H.; Chen, I.-M.; Wilson, C.S.; Bedrick, E.J.; Harvey, R.C.; Atlas, S.R.; Devidas, M.; Mullighan, C.G.; Wang, X.; Murphy, M.; et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood 2010, 115, 1394–1405.
- Frank, O.; Brors, B.; Fabarius, A.; Li, L.; Haak, M.; Merk, S.; Schwindel, U.; Zheng, C.; Müller, M.C.; Gretz, N.; et al. Gene expression signature of primary imatinib-resistant chronic myeloid leukemia patients. Leukemia 2006, 20, 1400–1407.
- Pise-Masison, C.A.; Radonovich, M.; Mahieux, R.; Chatterjee, P.; Whiteford, C.; Duvall, J.; Guillerm, C.; Gessain, A.; Brady, J.N. Transcription profile of cells infected with human T-cell leukemia virus type I compared with activated lymphocytes. Cancer Res. 2002, 62, 3562–3571.
- Nagarajan, A.; Dogra, S.K.; Sun, L.; Gandotra, N.; Ho, T.; Cai, G.; Cline, G.; Kumar, P.; Cowles, R.A.; Wajapeyee, N. Paraoxonase 2 Facilitates Pancreatic Cancer Growth and Metastasis by Stimulating GLUT1-Mediated Glucose Transport. Mol. Cell 2017, 67, 685–701.e6.
- Bacchetti, T.; Sartini, D.; Pozzi, V.; Cacciamani, T.; Ferretti, G.; Emanuelli, M. Exploring the role of Paraoxonase-2 in bladder cancer: Analyses performed on tissue samples, urines and cell culturess. Oncotarget 2017, 8, 28785–28795.
- Tseng, J.-H.; Chen, C.-Y.; Chen, P.-C.; Hsiao, S.-H.; Fan, C.-C.; Liang, Y.-C.; Chen, C.-P. Valproic acid inhibits glioblastoma multiforme cell growth via paraoxonase 2 expression. Oncotarget 2017, 8, 14666–14679.
- Krüger, M.; Amort, J.; Wilgenbus, P.; Helmstädter, J.P.; Grechowa, I.; Ebert, J.; Tenzer, S.; Moergel, M.; Witte, I.; Horke, S. The anti-apoptotic PON2 protein is Wnt/β-catenin-regulated and correlates with radiotherapy resistance in OSCC patients. Oncotarget 2016, 7, 51082–51095.
- Rosenblat, M.; Draganov, D.; Watson, C.E.; Bisgaier, C.L.; La Du, B.N.; Aviram, M. Mouse Macrophage Paraoxonase 2 Activity Is Increased Whereas Cellular Paraoxonase 3 Activity Is Decreased Under Oxidative Stress. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 468–474.
- Fuhrman, B.; Gantman, A.; Khateeb, J.; Volkova, N.; Horke, S.; Kiyan, J.; Dumler, I.; Aviram, M. Urokinase activates macrophage PON2 gene transcription via the PI3K/ROS/MEK/SREBP-2 signalling cascade mediated by the PDGFR-β. Cardiovasc. Res. 2009, 84, 145–154.
- Shiner, M.; Fuhrman, B.; Aviram, M. Paraoxonase 2 (PON2) expression is upregulated via a reduced-nicotinamide-adenine-dinucleotide-phosphate (NADPH)-oxidase-dependent mechanism during monocytes differentiation into macrophages. Free Radic. Biol. Med. 2004, 37, 2052–2063.
- Azizi, F.; Rahmani, M.; Raiszadeh, F.; Solati, M.; Navab, M. Association of lipids, lipoproteins, apolipoproteins and paraoxonase enzyme activity with premature coronary artery disease. Coron. Artery Dis. 2002, 13, 9–16.
- Schweikert, E.-M.; Devarajan, A.; Witte, I.; Wilgenbus, P.; Amort, J.; Förstermann, U.; Shabazian, A.; Grijalva, V.; Shih, D.M.; Farias-Eisner, R.; et al. PON3 is upregulated in cancer tissues and protects against mitochondrial superoxide-mediated cell death. Cell Death Differ. 2012, 19, 1549–1560.
- Krüger, M.; Pabst, A.; Al-Nawas, B.; Horke, S.; Moergel, M. Paraoxonase-2 (PON2) protects oral squamous cell cancer cells against irradiation-induced apoptosis. J. Cancer Res. Clin. Oncol. 2015, 141, 1757–1766.
- Devarajan, A.; Su, F.; Grijalva, V.; Yalamanchi, M.; Yalamanchi, A.; Gao, F.; Trost, H.; Nwokedi, J.; Farias-Eisner, G.; Farias-Eisner, R.; et al. Paraoxonase 2 overexpression inhibits tumor development in a mouse model of ovarian cancer. Cell Death Dis. 2018, 9, 1–16.
- Klover, P.J.; Zimmers, T.A.; Koniaris, L.G.; Mooney, R.A. Chronic Exposure to Interleukin-6 Causes Hepatic Insulin Resistance in Mice. Diabetes 2003, 52, 2784–2789.
- Shoelson, S.E.; Lee, J.; Goldfine, A.B. Inflammation and insulin resistance. J. Clin. Investig. 2006, 116, 1793–1801.
- Cai, D.; Yuan, M.; Frantz, D.F.; Melendez, P.A.; Hansen, L.; Lee, J.; Shoelson, S.E. Local and systemic insulin resistance resulting from hepatic activation of IKK-β and NF-κB. Nat. Med. 2005, 11, 183–190.
- Bourquard, N.; Ng, C.J.; Reddy, S.T. Impaired hepatic insulin signalling in PON2-deficient mice: A novel role for the PON2/apoE axis on the macrophage inflammatory response. Biochem. J. 2011, 436, 91–100.
- Song, B.; Scheuner, D.; Ron, D.; Pennathur, S.; Kaufman, R.J. Chop deletion reduces oxidative stress, improves β cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Investig. 2008, 118, 3378–3389.
- Giordano, G.; Cole, T.B.; Furlong, C.E.; Costa, L.G. Paraoxonase 2 (PON2) in the mouse central nervous system: A neuroprotective role? Toxicol. Appl. Pharmacol. 2011, 256, 369–378.
- Rosenblat, M.; Coleman, R.; Reddy, S.T.; Aviram, M. Paraoxonase 2 attenuates macrophage triglyceride accumulation via inhibition of diacylglycerol acyltransferase 1. J. Lipid Res. 2009, 50, 870–879.
- Higgins, G.C.; Beart, P.M.; Shin, Y.S.; Chen, M.J.; Cheung, N.S.; Nagley, P. Oxidative Stress: Emerging Mitochondrial and Cellular Themes and Variations in Neuronal Injury. J. Alzheimers Dis. 2010, 20, S453–S473.
- Mackness, B.; Beltran-Debon, R.; Aragones, G.; Joven, J.; Camps, J.; Mackness, M. Human tissue distribution of paraoxonases 1 and 2 mRNA. IUBMB Life 2010, 62, 480–482.
- Costa, L.G.; De Laat, R.; Dao, K.; Pellacani, C.; Cole, T.B.; Furlong, C.E. Paraoxonase-2 (PON2) in brain and its potential role in neuroprotection. Neurotoxicology 2014, 43, 3–9.
- Costa, L.G.; Tait, L.; De Laat, R.; Dao, K.; Giordano, G.; Pellacani, C.; Cole, T.B.; Furlong, C.E. Modulation of Paraoxonase 2 (PON2) in Mouse Brain by the Polyphenol Quercetin: A Mechanism of Neuroprotection? Neurochem. Res. 2013, 38, 1809–1818.
- Horke, S.; Witte, I.; Altenhöfer, S.; Wilgenbus, P.; Goldeck, M.; Förstermann, U.; Xiao, J.; Kramer, G.L.; Haines, D.C.; Chowdhary, P.K.; et al. Paraoxonase 2 is down-regulated by the Pseudomonas aeruginosa quorumsensing signal N-(3-oxododecanoyl)-L-homoserine lactone and attenuates oxidative stress induced by pyocyanin. Biochem. J. 2010, 426, 73–83.
- Shamir, R.; Hartman, C.; Karry, R.; Pavlotzky, E.; Eliakim, R.; Lachter, J.; Suissa, A.; Aviram, M. Paraoxonases (PONs) 1, 2, and 3 are expressed in human and mouse gastrointestinal tract and in Caco-2 cell line: Selective secretion of PON1 and PON2. Free Radic. Biol. Med. 2005, 39, 336–344.
- Levy, E.; Trudel, K.; Bendayan, M.; Seidman, E.; Delvin, E.; Elchebly, M.; Lavoie, J.-C.; Precourt, L.-P.; Amre, D.; Sinnett, D. Biological role, protein expression, subcellular localization, and oxidative stress response of paraoxonase 2 in the intestine of humans and rats. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G1252–G1261.
- Precourt, L.-P.; Seidman, E.; Delvin, E.; Amre, D.; Deslandres, C.; Dominguez, M.; Sinnett, D.; Levy, E. Comparative expression analysis reveals differences in the regulation of intestinal paraoxonase family members. Int. J. Biochem. Cell Biol. 2009, 41, 1628–1637.
- Hegele, R.A. Paraoxonase genes and disease. Ann. Med. 1999, 31, 217–224.
- Nie, Y.; Luo, D.; Yang, M.; Wang, Y.; Xiong, L.; Gao, L.; Liu, Y.; Liu, H. A Meta-Analysis on the Relationship of the PON Genes and Alzheimer Disease. J. Geriatr. Psychiatry Neurol. 2017, 30, 303–310.
- LeDuc, V.; Legault, V.; Dea, D.; Poirier, J. Normalization of gene expression using SYBR green qPCR: A case for paraoxonase 1 and 2 in Alzheimer’s disease brains. J. Neurosci. Methods 2011, 200, 14–19.
- Mu, N.; Xu, S.C.; Chang, Q.; Rao, D.P.; Chen, J.P.; Ma, C. Study of lipids, insulin metabolism, and paraoxonase-2-311 polymorphism in patients with different subtypes of Alzheimer’s disease (translated version). East Asian Arch. Psychiatry 2013, 23, 114–119.
- Janka, Z.; Juhász, A.; Rimanóczy, Á.; Boda, K.; Márki-Zay, J.; Kálmán, J. Codon 311 (Cys → Ser) polymorphism of paraoxonase-2 gene is associated with apolipoprotein E4 allele in both Alzheimer’s and vascular dementias. Mol. Psychiatry 2002, 7, 110–112.
- Shi, J.; Zhang, S.; Tang, M.; Liu, X.; Li, T.; Han, H.; Wang, Y.; Guo, Y.; Zhao, J.; Li, H.; et al. Possible association between Cys311Ser polymorphism of paraoxonase 2 gene and late-onset Alzheimer’s disease in Chinese. Mol. Brain Res. 2004, 120, 201–204.
- Erlich, P.M.; Lunetta, K.L.; Cupples, L.A.; Huyck, M.; Green, R.C.; Baldwin, C.T.; Farrer, L.A.; Auerbach, S.; Akomolafe, A.; Griffith, P.; et al. Polymorphisms in the PON gene cluster are associated with Alzheimer disease. Hum. Mol. Genet. 2006, 15, 77–85.
- Akhmedova, S.N.; Yakimovsky, A.K.; Schwartz, E.I. Paraoxonase 1 Met–Leu 54 polymorphism is associated with Parkinson’s disease. J. Neurol. Sci. 2001, 184, 179–182.
- Carmine, A.; Buervenich, S.; Sydow, O.; Anvret, M.; Olson, L. Further evidence for an association of the Paraoxonase 1 (PON1) Met-54 allele with Parkinson’s disease. Mov. Disord. 2002, 17, 764–766.
- Hirsch, E.C.; Graybiel, A.M.; Agid, Y.A. Melanized dopaminergic neurons are differentially susceptible to degeneration in Parkinson’s disease. Nat. Cell Biol. 1988, 334, 345–348.
- Tanner, C.M. Parkinson Disease in TwinsAn Etiologic Study. JAMA 1999, 281, 341–346.
- Polymeropoulos, M.H. Mutation in the -Synuclein Gene Identified in Families with Parkinson’s Disease. Science 1997, 276, 2045–2047.
- Kitada, T.; Asakawa, S.; Hattori, N.; Matsumine, H.; Yamamura, Y.; Minoshima, S.; Yokochi, M.; Mizuno, Y.; Shimizu, N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nat. Cell Biol. 1998, 392, 605–608.
- Leroy, E.; Boyer, R.; Auburger, G.; Leube, B.; Ulm, G.; Mezey, E.; Harta, G.; Brownstein, M.J.; Jonnalagada, S.; Chernova, T.; et al. The ubiquitin pathway in Parkinson’s disease. Nat. Cell Biol. 1998, 395, 451–452.
- Krüger, R.; Kuhn, W.; Müller, T.; Woitalla, D.; Graeber, M.B.; Kösel, S.; Przuntek, H.; Epplen, J.T.; Schols, L.; Riess, O. AlaSOPro mutation in the gene encoding α-synuclein in Parkinson’s disease. Nat. Genet. 1998, 18, 106–108.
- Bonifati, V.; Rizzu, P.; Van Baren, M.J.; Schaap, O.; Breedveld, G.J.; Krieger, E.; Dekker, M.C.J.; Squitieri, F.; Ibanez, P.; Joosse, M.; et al. Mutations in the DJ-1 Gene Associated with Autosomal Recessive Early-Onset Parkinsonism. Science 2002, 299, 256–259.
- Ewing, R.M.; Chu, P.; Elisma, F.; Li, H.; Taylor, P.; Climie, S.; McBroom-Cerajewski, L.; Robinson, M.D.; O’Connor, L.; Li, M.; et al. Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol. Syst. Biol. 2007, 3, 89.
- Parsanejad, M.; Bourquard, N.; Qu, D.; Zhang, Y.; Huang, E.; Rousseaux, M.W.C.; Aleyasin, H.; Irrcher, I.; Callaghan, S.; Vaillant, D.C.; et al. DJ-1 Interacts with and Regulates Paraoxonase-2, an Enzyme Critical for Neuronal Survival in Response to Oxidative Stress. PLoS ONE 2014, 9, e106601.
- Yang, J.; Kim, K.S.; Iyirhiaro, G.O.; Marcogliese, P.C.; Callaghan, S.M.; Qu, D.; Kim, W.J.; Slack, R.S.; Park, D.S. DJ-1 modulates the unfolded protein response and cell death via upregulation of ATF4 following ER stress. Cell Death Dis. 2019, 10, 135.
- Holtz, W.A.; O’Malley, K.L. Parkinsonian Mimetics Induce Aspects of Unfolded Protein Response in Death of Dopaminergic Neurons. J. Biol. Chem. 2003, 278, 19367–19377.
- Aguirre-Vidal, Y.; Morales-Montor, J.; De León, C.T.G.; Ostoa-Saloma, P.; Díaz-Zaragoza, M.; Montes, S.; Arteaga-Silva, M.; Antonio, M.-N. Protection induced by estradiol benzoate in the MPP+ rat model of Parkinson’s disease is associated with the regulation of the inflammatory cytokine profile in the nigro striatum. J. Neuroimmunol. 2020, 349, 577426.
- Reichert, C.O.; Levy, D.; Bydlowski, S.P. Paraoxonase Role in Human Neurodegenerative Diseases. Antioxidants 2020, 10, 11.
- Narain, P.; Padhi, A.K.; Dave, U.; Mishra, D.; Bhatia, R.; Vivekanandan, P.; Gomes, J. Identification and characterization of novel and rare susceptible variants in Indian amyotrophic lateral sclerosis patients. Neurogenetics 2019, 20, 197–208.
- Obrador, E.; Salvador, R.; López-Blanch, R.; Jihad-Jebbar, A.; Vallés, S.L.; Estrela, J.M. Oxidative Stress, Neuroinflammation and Mitochondria in the Pathophysiology of Amyotrophic Lateral Sclerosis. Antioxidants 2020, 9, 901.
- Herrando-Grabulosa, M.; Gaja-Capdevila, N.; Vela, J.M.; Navarro, X. Sigma 1 receptor as a therapeutic target for amyotrophic lateral sclerosis. Br. J. Pharmacol. 2020, 72, 1–17.
- Saeed, M.; Siddique, N.; Hung, W.Y.; Usacheva, E.; Liu, E.; Sufit, R.L.; Heller, S.L.; Haines, J.L.; Pericak-Vance, M.; Siddique, T. Paraoxonase cluster polymorphisms are associated with sporadic ALS. Neurology 2006, 67, 771–776.
- Slowik, A.; Tomik, B.; Wolkow, P.P.; Partyka, D.; Turaj, W.; Malecki, M.T.; Pera, J.; Dziedzic, T.; Szczudlik, A.; Figlewicz, D.A. Paraoxonase gene polymorphisms and sporadic ALS. Neurology 2006, 67, 766–770.
- Landers, J.E.; Shi, L.; Cho, T.-J.; Glass, J.D.; Shaw, P.; Leigh, P.N.; Diekstra, F.; Polak, M.; Rodriguez-Leyva, I.; Niemann, S.; et al. A common haplotype within the PON1 promoter region is associated with sporadic ALS. Amyotroph. Lateral Scler. 2008, 9, 306–314.
- Valdmanis, P.N.; Kabashi, E.; Dyck, A.; Hince, P.; Lee, J.; Dion, P.; D’Amour, M.; Souchon, F.; Bouchard, J.-P.; Salachas, F.; et al. Association of paraoxonase gene cluster polymorphisms with ALS in France, Quebec, and Sweden. Neurology 2008, 71, 514–520.
- Ticozzi, N.; Ba, A.L.L.; Bs, P.J.K.; Glass, J.D.; Wills, A.-M.; Van Blitterswijk, M.; Bosco, D.A.; Rodriguez-Leyva, I.; Gellera, C.; Ratti, A.; et al. Paraoxonase gene mutations in amyotrophic lateral sclerosis. Ann. Neurol. 2010, 68, 102–107.
- Ricci, C.; Battistini, S.; Cozzi, L.; Benigni, M.; Origone, P.; Verriello, L.; Lunetta, C.; Cereda, C.; Milani, P.; Greco, G.; et al. Lack of association of PON polymorphisms with sporadic ALS in an Italian population. Neurobiol. Aging 2011, 32, 552.e7–552.e13.
- Chen, Y.; Huang, R.; Chen, K.; Song, W.; Yang, Y.; Zhao, B.; Li, J.; Shang, H.-F. Association analysis of PON polymorphisms in sporadic ALS in a Chinese population. Neurobiol. Aging 2012, 33, 2949.e1–2949.e3.
- Gagliardi, S.; Abel, K.; Bianchi, M.; Milani, P.; Bernuzzi, S.; Corato, M.; Ceroni, M.; Cashman, J.R.; Cereda, C. Regulation of FMO and PON Detoxication Systems in ALS Human Tissues. Neurotox. Res. 2012, 23, 370–377.
- Wheeler, J.G.; Keavney, B.D.; Watkins, H.; Collins, R.; Danesh, J. Four paraoxonase gene polymorphisms in 11 212 cases of coronary heart disease and 12 786 controls: Meta-analysis of 43 studies. Lancet 2004, 363, 689–695.
- Liu, D.; Yang, J.; Feng, B.; Lu, W.; Zhao, C.; Li, L. Mendelian randomization analysis identified genes pleiotropically associated with the risk and prognosis of COVID-19. J. Infect. 2021, 82, 126–132.
- Rodríguez-Esparragón, F.; Fernández, J.C.L.; Ríos, N.B.; García-Bello, M.A.; Velazquez, E.H.; Cappiello, L.; Pérez, J.C.R. Paraoxonase 1 and 2 gene variants and the ischemic stroke risk in Gran Canaria population: An association study and meta-analysis. Int. J. Neurosci. 2016, 127, 191–198.
- Wu, S.S.; Yu, J.N.; Jiao, J.; Chen, G.S.; Zhang, C.Y.; Yu, S.F. Association between PON2 gene polymorphisms and susceptibility to noise-induced hearing loss. Chin. J. Ind. Hyg. Occup. Dis. 2020, 38, 128–132.
- Li, X.; Cao, J.; Wang, J.; Song, H.; Ji, G.; Dong, Q.; Wei, C.; Cao, Y.; Wang, B.; Zhu, B.; et al. PON2 and ATP2B2 gene polymorphisms with noise-induced hearing loss. J. Thorac. Dis. 2016, 8, 430–438.




