
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ugur Gezer | -- | 1856 | 2022-06-15 10:37:21 | | | |
| 2 | Peter Tang | + 10 word(s) | 1866 | 2022-06-15 11:46:12 | | |
Video Upload Options
Liquid biopsy is a broad term that refers to the testing of body fluids for biomarkers that correlate with a pathological condition. While a variety of body-fluid components (e.g., circulating tumor cells, extracellular vesicles, RNA, proteins, and metabolites) are studied as potential liquid biopsy biomarkers, cell-free DNA (cfDNA) has attracted the most attention. The total cfDNA population in a typical biospecimen represents an immensely rich source of biological and pathological information and has demonstrated significant potential as a versatile biomarker in oncology, non-invasive prenatal testing, and transplant monitoring. As a significant portion of cfDNA is composed of repeat DNA sequences and some families (e.g., pericentric satellites) were recently shown to be overrepresented in cfDNA populations vs their genomic abundance, it holds great potential for developing liquid biopsy-based biomarkers for the early detection and management of patients with cancer.
1. Introduction
2. Repeat DNA Content of the Human Genome and Cell-Free DNA
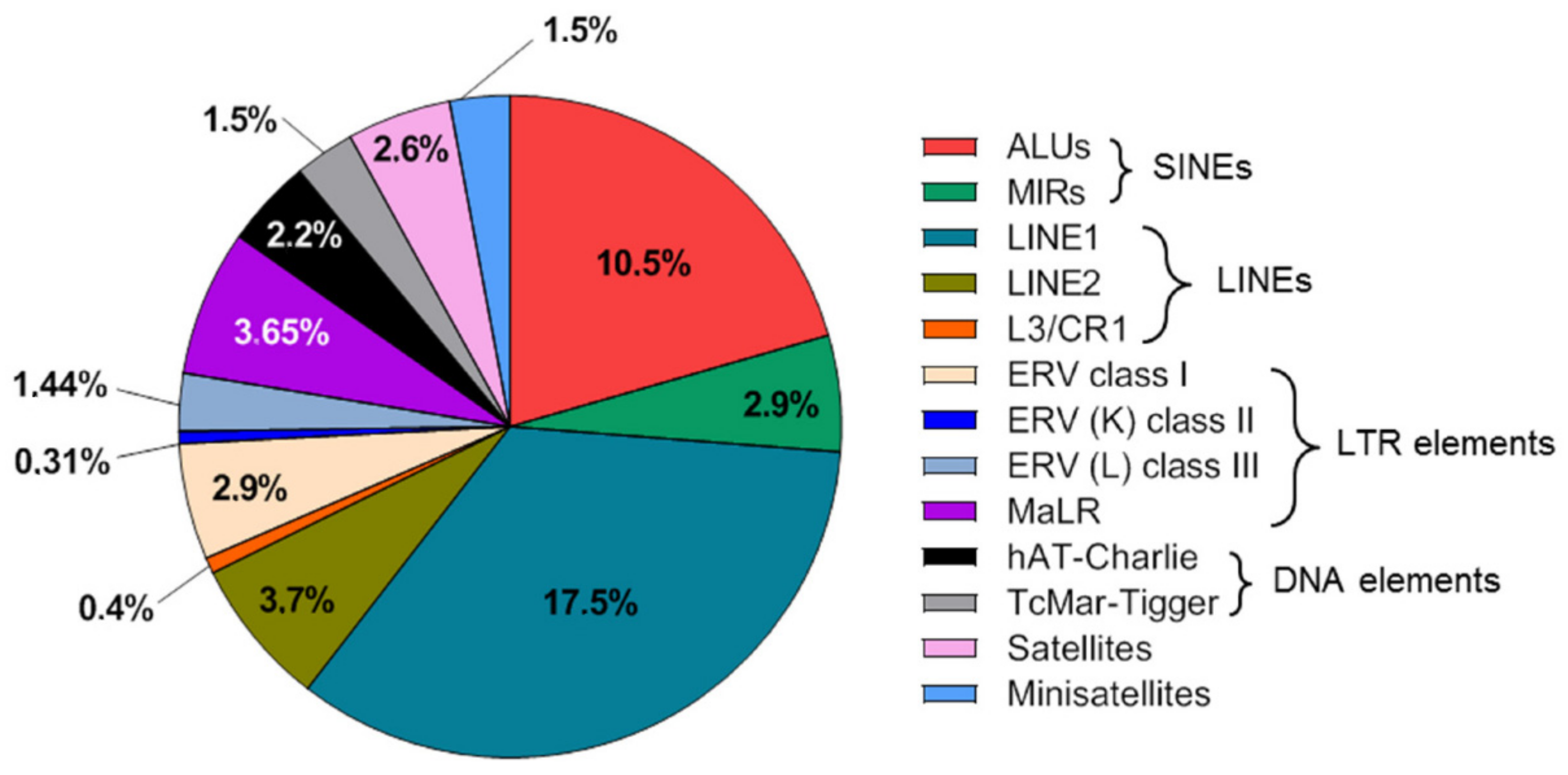
3. Application of Cell-Free Repetitive DNA in Liquid Biopsies of Cancer
Table 1 summarizes applications of repetitive DNA elements in liquid biopsy.
Table 1. An overview on applications of repetitive DNA elements in liquid biopsy.
|
Repeat Name |
Repeat Type |
Liquid Biopsy Application in Patients with Cancer |
References |
|---|---|---|---|
|
LINE1 |
Interspersed |
Quantification of total cfDNA |
|
|
Assessment of cfDNA integrity |
[40] |
||
|
Detection of aneuploidy by amplicon sequencing |
|||
|
Assessment of cfDNA global methylation status |
|||
|
ALU |
Interspersed |
Quantification of total cfDNA |
|
|
Assessment of cfDNA integrity |
[39][57][58][61][62][64][65][66][67][68][69][70][71][72][73][74] |
||
|
Detection of aneuploidy by amplicon sequencing |
[43] |
||
|
Human satellite 2 |
Tandem |
Quantification of total cfDNA |
[75] |
|
Microsatellites |
Tandem |
Assessment of microsatellite instability via cfDNA |
|
|
Telomeric repeats |
Tandem |
Quantification of telomeric cfDNA |
|
|
Assessment of telomere length in cf DNA |
Table 2 summarizes studies that utilized cell-free repetitive DNA in clinical use of cancer patients.
Table 2. Studies that utilized cell-free repetitive DNA in clinical use of cancer patients.
|
Clinical Use |
Repeat Type |
References |
|---|---|---|
|
Evaluation of diagnostic potential of cedant |
LINE1 |
|
|
ALU |
[39][44][56][57][58][59][60][61][62][63][64][65][66][67][69][70][71][72][74][87][88] |
|
|
Human satellite 2 |
[75] |
|
|
Microsatellites |
||
|
Prognostic significance of cfDNA |
LINE1 |
|
|
ALU |
||
|
Microsatellites |
[78] |
|
|
Predicting the response to neoadjuvant chemotherapy |
ALU |
|
|
Monitoring of cancer patients |
LINE1 |
|
|
ALU |
||
|
Microsatellites |
References
- González-Masiá, J.A.; García-Olmo, D.; García-Olmo, D.C. Circulating nucleic acids in plasma and serum (CNAPS): Applications in oncology. OncoTargets Ther. 2013, 6, 819–832.
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2018, 20, 71–88.
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019, 17, 100087.
- Bronkhorst, A.J.; Ungerer, V.; Diehl, F.; Anker, P.; Dor, Y.; Fleischhacker, M.; Gahan, P.B.; Hui, L.; Holdenrieder, S.; Thierry, A.R. Towards systematic nomenclature for cell-free DNA. Hum. Genet. 2020, 140, 565–578.
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic Characteristics of the DNA Found in the Plasma of Cancer Patients. Oncology 1989, 46, 318–322.
- Sidransky, D.; Von Eschenbach, A.; Tsai, Y.C.; Jones, P.; Summerhayes, I.; Marshall, F.; Paul, M.; Green, P.; Hamilton, S.R.; Frost, P.; et al. Identification of p53 gene mutations in bladder cancers and urine samples. Science 1991, 252, 706–709.
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484.
- Jin, J.; Wu, X.; Yin, J.; Li, M.; Shen, J.; Li, J.; Zhao, Y.; Zhao, Q.; Wu, J.; Wen, Q.; et al. Identification of Genetic Mutations in Cancer: Challenge and Opportunity in the New Era of Targeted Therapy. Front. Oncol. 2019, 9, 263.
- Cisneros-Villanueva, M.; Hidalgo-Pérez, L.; Rios-Romero, M.; Cedro-Tanda, A.; Ruiz-Villavicencio, C.A.; Page, K.; Hastings, R.; Fernandez-Garcia, D.; Allsopp, R.; Fonseca-Montaño, M.A.; et al. Cell-free DNA analysis in current cancer clinical trials: A review. Br. J. Cancer 2022, 126, 391–400.
- Aucamp, J.; Bronkhorst, A.J.; Badenhorst, C.P.S.; Pretorius, P.J. The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature. Biol. Rev. 2018, 93, 1649–1683.
- Suzuki, N.; Kamataki, A.; Yamaki, J.; Homma, Y. Characterization of circulating DNA in healthy human plasma. Clin. Chim. Acta 2008, 387, 55–58.
- Van der Vaart, M.; Pretorius, P.J. Characterization of circulating DNA in healthy human plasma. Clin. Chim. Acta Int. J. Clin. Chem. 2008, 395, 186.
- Lo, Y.D.; Chiu, R.W. Next-Generation Sequencing of Plasma/Serum DNA: An Emerging Research and Molecular Diagnostic Tool. Clin. Chem. 2009, 55, 607–608.
- Grabuschnig, S.; Bronkhorst, A.J.; Holdenrieder, S.; Rodriguez, I.R.; Schliep, K.P.; Schwendenwein, D.; Ungerer, V.; Sensen, C.W. Putative Origins of Cell-Free DNA in Humans: A Review of Active and Passive Nucleic Acid Release Mechanisms. Int. J. Mol. Sci. 2020, 21, 8062.
- Alborelli, I.; Generali, D.; Jermann, P.; Cappelletti, M.R.; Ferrero, G.; Scaggiante, B.; Bortul, M.; Zanconati, F.; Nicolet, S.; Haegele, J.; et al. Cell-free DNA analysis in healthy individuals by next-generation sequencing: A proof of concept and technical validation study. Cell Death Dis. 2019, 10, 534.
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24.
- Aravanis, A.M.; Lee, M.; Klausner, R.D. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 2017, 168, 571–574.
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930.
- Ding, S.C.; Lo, Y.D. Cell-Free DNA Fragmentomics in Liquid Biopsy. Diagnostics 2022, 12, 978.
- Van der Pol, Y.; Mouliere, F. Toward the Early Detection of Cancer by Decoding the Epigenetic and Environmental Fingerprints of Cell-Free DNA. Cancer Cell 2019, 36, 350–368.
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. Early detection of cancer using circulating tumor DNA: Biological, physiological and analytical considerations. Crit. Rev. Clin. Lab. Sci. 2019, 57, 253–269.
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921.
- De Koning, A.P.J.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive Elements May Comprise Over Two-Thirds of the Human Genome. PLoS Genet. 2011, 7, e1002384.
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.e3.
- Bois, P.R. Hypermutable minisatellites, a human affair? Genomics 2003, 81, 349–355.
- Pezer, Z.; Brajkovic, J.; Feliciello, I.; Ugarković, D. Satellite DNA-Mediated Effects on Genome Regulation. Repetitive DNA 2012, 7, 153–169.
- Hall, L.L.; Byron, M.; Carone, D.M.; Whitfield, T.W.; Pouliot, G.P.; Fischer, A.; Jones, P.; Lawrence, J.B. Demethylated HSATII DNA and HSATII RNA Foci Sequester PRC1 and MeCP2 into Cancer-Specific Nuclear Bodies. Cell Rep. 2017, 18, 2943–2956.
- Bersani, F.; Lee, E.; Kharchenko, P.V.; Xu, A.W.; Liu, M.; Xega, K.; MacKenzie, O.C.; Brannigan, B.W.; Wittner, B.S.; Jung, H.; et al. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. USA 2015, 112, 15148–15153.
- Criscione, S.W.; Zhang, Y.; Thompson, W.; Sedivy, J.M.; Neretti, N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genom. 2014, 15, 583.
- Levin, H.L.; Moran, J.V. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 2011, 12, 615–627.
- Beck, J.; Urnovitz, H.B.; Riggert, J.; Clerici, M.; Schütz, E. Profile of the Circulating DNA in Apparently Healthy Individuals. Clin. Chem. 2009, 55, 730–738.
- Van der Vaart, M.; Semenov, D.V.; Kuligina, E.V.; Richter, V.A.; Pretorius, P.J. Characterisation of circulating DNA by parallel tagged sequencing on the 454 platform. Clin. Chim. Acta 2009, 409, 21–27.
- Bronkhorst, A.J.; Wentzel, J.F.; Aucamp, J.; van Dyk, E.; du Plessis, L.; Pretorius, P.J. Characterization of the cell-free DNA released by cultured cancer cells. Biochim. Biophys. Acta 2016, 1863, 157–165.
- Bronkhorst, A.J.; Wentzel, J.F.; Ungerer, V.; Peters, D.L.; Aucamp, J.; de Villiers, E.P.; Holdenrieder, S.; Pretorius, P.J. Sequence analysis of cell-free DNA derived from cultured human bone osteosarcoma (143B) cells. Tumour Biol. 2018, 40, 1010428318801190.
- Grabuschnig, S.; Soh, J.; Heidinger, P.; Bachler, T.; Hirschböck, E.; Rodriguez, I.R.; Schwendenwein, D.; Sensen, C.W. Circulating cell-free DNA is predominantly composed of retrotransposable elements and non-telomeric satellite DNA. J. Biotechnol. 2020, 313, 48–56.
- Sunami, E.; Vu, A.-T.; Nguyen, S.L.; Giuliano, A.E.; Hoon, D.S.B. Quantification of LINE1 in Circulating DNA as a Molecular Biomarker of Breast Cancer. Ann. N. Y. Acad. Sci. 2008, 1137, 171–174.
- Wei, L.; Wu, W.; Han, L.; Yu, W.; Du, Y. A quantitative analysis of the potential biomarkers of non-small cell lung cancer by circulating cell-free DNA. Oncol. Lett. 2018, 16, 4353–4360.
- Wang, X.; Wang, L.; Su, Y.; Yue, Z.; Xing, T.; Zhao, W.; Zhao, Q.; Duan, C.; Huang, C.; Zhang, D.; et al. Plasma cell-free DNA quantification is highly correlated to tumor burden in children with neuroblastoma. Cancer Med. 2018, 7, 3022–3030.
- Madhavan, D.; Wallwiener, M.; Bents, K.; Zucknick, M.; Nees, J.; Schott, S.; Cuk, K.; Riethdorf, S.; Trumpp, A.; Pantel, K.; et al. Plasma DNA integrity as a biomarker for primary and metastatic breast cancer and potential marker for early diagnosis. Breast Cancer Res. Treat. 2014, 146, 163–174.
- Cheng, J.; Cuk, K.; Heil, J.; Golatta, M.; Schott, S.; Sohn, C.; Schneeweiss, A.; Burwinkel, B.; Surowy, H. Cell-free circulating DNA integrity is an independent predictor of impending breast cancer recurrence. Oncotarget 2017, 8, 54537–54547.
- Kinde, I.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. FAST-SeqS: A Simple and Efficient Method for the Detection of Aneuploidy by Massively Parallel Sequencing. PLoS ONE 2012, 7, e41162.
- Douville, C.; Springer, S.; Kinde, I.; Cohen, J.D.; Hruban, R.H.; Lennon, A.M.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B.; Karchin, R. Detection of aneuploidy in patients with cancer through amplification of long interspersed nucleotide elements (LINEs). Proc. Natl. Acad. Sci. USA 2018, 115, 1871–1876.
- Douville, C.; Cohen, J.D.; Ptak, J.; Popoli, M.; Schaefer, J.; Silliman, N.; Dobbyn, L.; Schoen, R.E.; Tie, J.; Gibbs, P.; et al. Assessing aneuploidy with repetitive element sequencing. Proc. Natl. Acad. Sci. USA 2020, 117, 4858–4863.
- Park, M.-K.; Lee, J.-C.; Lee, J.-W.; Hwang, S.-J. Alu cell-free DNA concentration, Alu index, and LINE-1 hypomethylation as a cancer predictor. Clin. Biochem. 2021, 94, 67–73.
- Ko, K.; Kananazawa, Y.; Yamada, T.; Kakinuma, D.; Matsuno, K.; Ando, F.; Kuriyama, S.; Matsuda, A.; Yoshida, H. Methylation status and long-fragment cell-free DNA are prognostic biomarkers for gastric cancer. Cancer Med. 2021, 10, 2003–2012.
- Nagai, Y.; Sunami, E.; Yamamoto, Y.; Hata, K.; Okada, S.; Murono, K.; Yasuda, K.; Otani, K.; Nishikawa, T.; Tanaka, T.; et al. LINE-1 hypomethylation status of circulating cell-free DNA in plasma as a biomarker for colorectal cancer. Oncotarget 2017, 8, 11906–11916.
- Lee, K.-H.; Shin, T.-J.; Kim, W.-H.; Cho, J.-Y. Methylation of LINE-1 in cell-free DNA serves as a liquid biopsy biomarker for human breast cancers and dog mammary tumors. Sci. Rep. 2019, 9, 175.
- Kaneko, M.; Kotake, M.; Bando, H.; Yamada, T.; Takemura, H.; Minamoto, T. Prognostic and predictive significance of long interspersed nucleotide element-1 methylation in advanced-stage colorectal cancer. BMC Cancer 2016, 16, 945.
- Tangkijvanich, P.; Hourpai, N.; Rattanatanyong, P.; Wisedopas, N.; Mahachai, V.; Mutirangura, A. Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clin. Chim. Acta 2007, 379, 127–133.
- Boldrin, E.; Curtarello, M.; Dallan, M.; Alfieri, R.; Realdon, S.; Fassan, M.; Saggioro, D. Detection of LINE-1 hypomethylation in cfDNA of Esophageal Adenocarcinoma Patients. Int. J. Mol. Sci. 2020, 21, 1547.
- Misawa, K.; Yamada, S.; Mima, M.; Nakagawa, T.; Kurokawa, T.; Imai, A.; Mochizuki, D.; Shinmura, D.; Yamada, T.; Kita, J.; et al. Long interspersed nuclear element 1 hypomethylation has novel prognostic value and potential utility in liquid biopsy for oral cavity cancer. Biomark. Res. 2020, 8, 53.
- Hoshimoto, S.; Kuo, C.T.; Chong, K.K.; Takeshima, T.-L.; Takei, Y.; Li, M.W.; Huang, S.K.; Sim, M.-S.; Morton, D.L.; Hoon, D.S. AIM1 and LINE-1 Epigenetic Aberrations in Tumor and Serum Relate to Melanoma Progression and Disease Outcome. J. Investig. Dermatol. 2012, 132, 1689–1697.
- Park, S.Y.; Seo, A.N.; Jung, H.Y.; Gwak, J.M.; Jung, N.; Cho, N.-Y.; Kang, G.H. Alu and LINE-1 Hypomethylation Is Associated with HER2 Enriched Subtype of Breast Cancer. PLoS ONE 2014, 9, e100429.
- Ramzy, I.I.; Omran, D.A.; Hamad, O.; Shaker, O.; Abboud, A. Evaluation of serum LINE-1 hypomethylation as a prognostic marker for hepatocellular carcinoma. Arab. J. Gastroenterol. 2011, 12, 139–142.
- Adusei, E.; Ahenkorah, J.; Adu-Aryee, N.A.; Adutwum-Ofosu, K.K.; Tagoe, E.A.; Koney, N.K.-K.; Nkansah, E.; Aryee, N.A.; Blay, R.M.; Hottor, B.A.; et al. Reduced Serum Circulation of Cell-Free DNA Following Chemotherapy in Breast Cancer Patients. Med. Sci. 2021, 9, 37.
- Leng, S.; Zheng, J.; Jin, Y.; Zhang, H.; Zhu, Y.; Wu, J.; Xu, Y.; Zhang, P. Plasma cell-free DNA level and its integrity as biomarkers to distinguish non-small cell lung cancer from tuberculosis. Clin. Chim. Acta 2018, 477, 160–165.
- Soliman, S.E.-S.; Alhanafy, A.M.; El-din Habib, M.S.; Hagag, M.; Ibrahem, R.A.L. Serum circulating cell free DNA as potential diagnostic and prognostic biomarker in non small cell lung cancer. Biochem. Biophys. Rep. 2018, 15, 45–51.
- Feng, J.; Gang, F.; Li, X.; Jin, T.; HouBao, H.; Yu, C.; Guorong, L. Plasma cell-free DNA and its DNA integrity as biomarker to distinguish prostate cancer from benign prostatic hyperplasia in patients with increased serum prostate-specific antigen. Int. Urol. Nephrol. 2013, 45, 1023–1028.
- Fawzy, A.; Sweify, K.M.; El-Fayoumy, H.M.; Nofal, N. Quantitative analysis of plasma cell-free DNA and its DNA integrity in patients with metastatic prostate cancer using ALU sequence. J. Egypt. Natl. Cancer Inst. 2016, 28, 235–242.
- Condappa, A.; McGrowder, D.; Aiken, W.; McLaughlin, W.; Gossell-Williams, M. Evaluation of Plasma Circulating Cell Free DNA Concentration and Integrity in Patients with Prostate Cancer in Jamaica: A Preliminary Study. Diseases 2020, 8, 34.
- Hao, T.B.; Shi, W.; Shen, X.J.; Qi, J.; Wu, X.H.; Wu, Y.; Tang, Y.Y.; Ju, S.Q. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br. J. Cancer 2014, 111, 1482–1489.
- Bedin, C.; Enzo, M.V.; Del Bianco, P.; Pucciarelli, S.; Nitti, D.; Agostini, M. Diagnostic and prognostic role of cell-free DNA testing for colorectal cancer patients. Int. J. Cancer 2017, 140, 1888–1898.
- Qian, C.; Ju, S.; Qi, J.; Zhao, J.; Shen, X.; Jing, R.; Yu, J.; Li, L.; Shi, Y.; Zhang, L.; et al. Alu-based cell-free DNA: A novel biomarker for screening of gastric cancer. Oncotarget 2017, 8, 54037–54045.
- Umetani, N.; Kim, J.; Hiramatsu, S.; Reber, H.A.; Hines, O.J.; Bilchik, A.J.; Hoon, D.S.B. Increased Integrity of Free Circulating DNA in Sera of Patients with Colorectal or Periampullary Cancer: Direct Quantitative PCR for ALU Repeats. Clin. Chem. 2006, 52, 1062–1069.
- Agostini, M.; Enzo, M.V.; Bedin, C.; Belardinelli, V.; Goldin, E.; Del Bianco, P.; Maschietto, E.; D’Angelo, E.; Izzi, L.; Saccani, A.; et al. Circulating cell-free DNA: A promising marker of regional lymphonode metastasis in breast cancer patients. Cancer Biomark. 2012, 11, 89–98.
- Hussein, N.A.; Mohamed, S.N.; Ahmed, M.A. Plasma ALU-247, ALU-115, and cfDNA Integrity as Diagnostic and Prognostic Biomarkers for Breast Cancer. Appl. Biochem. Biotechnol. 2019, 187, 1028–1045.
- Arko-Boham, B.; Aryee, N.A.; Blay, R.M.; Owusu, E.D.A.; Tagoe, E.A.; Shackie, E.-S.D.; Debrah, A.B.; Adu-Aryee, N.A. Circulating cell-free DNA integrity as a diagnostic and prognostic marker for breast and prostate cancers. Cancer Genet. 2019, 235, 65–71.
- Chudasama, D.Y.; Aladag, Z.; Felicien, M.I.; Hall, M.; Beeson, J.; Asadi, N.; Gidron, Y.; Karteris, E.; Anikin, V.B. Prognostic value of the DNA integrity index in patients with malignant lung tumors. Oncotarget 2018, 9, 21281–21288.
- Yörüker, E.E.; Özgür, E.; Keskin, M.; Dalay, N.; Holdenrieder, S.; Gezer, U. Assessment of circulating serum DNA integrity in colorectal cancer patients. Anticancer Res. 2015, 35, 2435–2440.
- Vizza, E.; Corrado, G.; de Angeli, M.; Carosi, M.; Mancini, E.; Baiocco, E.; Chiofalo, B.; Patrizi, L.; Zampa, A.; Piaggio, G.; et al. Serum DNA integrity index as a potential molecular biomarker in endometrial cancer. J. Exp. Clin. Cancer Res. 2018, 37, 16.
- Kumari, S.; Husain, N.; Agarwal, A.; Neyaz, A.; Gupta, S.; Chaturvedi, A.; Lohani, M.; Sonkar, A.A. Diagnostic Value of Circulating Free DNA Integrity and Global Methylation Status in Gall Bladder Carcinoma. Pathol. Oncol. Res. 2019, 25, 925–936.
- Zhang, R.; Pu, W.; Zhang, S.; Chen, L.; Zhu, W.; Xiao, L.; Xing, C.; Li, K. Clinical value of ALU concentration and integrity index for the early diagnosis of ovarian cancer: A retrospective cohort trial. PLoS ONE 2018, 13, e0191756.
- Nong, J.; Gong, Y.; Guan, Y.; Yi, X.; Yi, Y.; Chang, L.; Yang, L.; Lv, J.; Guo, Z.; Jia, H.; et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat. Commun. 2018, 9, 3114.
- Huang, A.; Zhang, X.; Zhou, S.-L.; Cao, Y.; Huang, X.-W.; Fan, J.; Yang, X.-R.; Zhou, J. Plasma Circulating Cell-free DNA Integrity as a Promising Biomarker for Diagnosis and Surveillance in Patients with Hepatocellular Carcinoma. J. Cancer 2016, 7, 1798–1803.
- Özgür, E.; Mayer, Z.; Keskin, M.; Yörüker, E.E.; Holdenrieder, S.; Gezer, U. Satellite 2 repeat DNA in blood plasma as a candidate biomarker for the detection of cancer. Clin. Chim. Acta 2021, 514, 74–79.
- Chen, X.Q.; Stroun, M.; Magnenat, J.-L.; Nicod, L.P.; Kurt, A.-M.; Lyautey, J.; Lederrey, C.; Anker, P. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat. Med. 1996, 2, 1033–1035.
- Nawroz, H.; Koch, W.M.; Anker, P.; Stroun, M.; Sidransky, D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat. Med. 1996, 2, 1035–1037.
- Gonzalez, R.; Silva, J.M.; Sanchez, A.; Dominguez, G.; Garcia, J.M.; Chen, X.Q.; Stroun, M.; Provencio, M.; España, P.; Anker, P.; et al. Microsatellite alterations and TP53 mutations in plasma DNA of small-cell lung cancer patients: Follow-up study and prognostic significance. Ann. Oncol. 2000, 11, 1097–1104.
- Mokarram, P.; Rismanchi, M.; Naeeni, M.A.; Samiee, S.M.; Paryan, M.; Alipour, A.; Honardar, Z.; Kavousipour, S.; Naghibalhossaini, F.; Mostafavi-Pour, Z.; et al. Microsatellite instability typing in serum and tissue of patients with colorectal cancer: Comparing real time PCR with hybridization probe and high-performance liquid chromatography. Mol. Biol. Rep. 2014, 41, 2835–2844.
- Silveira, A.B.; Bidard, F.-C.; Kasperek, A.; Melaabi, S.; Tanguy, M.-L.; Rodrigues, M.; Bataillon, G.; Cabel, L.; Buecher, B.; Pierga, J.-Y.; et al. High-Accuracy Determination of Microsatellite Instability Compatible with Liquid Biopsies. Clin. Chem. 2020, 66, 606–613.
- Boldrin, E.; Piano, M.A.; Alfieri, R.; Mazza, M.; Vassallo, L.; Scapinello, A.; Pilati, P.; Curtarello, M. MSI Analysis in Solid and Liquid Biopsies of Gastroesophageal Adenocarcinoma Patients: A Molecular Approach. Int. J. Mol. Sci. 2021, 22, 7244.
- Barata, P.; Agarwal, N.; Nussenzveig, R.; Gerendash, B.; Jaeger, E.; Hatton, W.; Ledet, E.; Lewis, B.; Layton, J.; Babiker, H.; et al. Clinical activity of pembrolizumab in metastatic prostate cancer with microsatellite instability high (MSI-H) detected by circulating tumor DNA. J. Immunother. Cancer 2020, 8, e001065.
- Wu, X.; Tanaka, H. Aberrant reduction of telomere repetitive sequences in plasma cell-free DNA for early breast cancer detection. Oncotarget 2015, 6, 29795–29807.
- Dey, S.; Marino, N.; Bishop, K.; Dahlgren, P.N.; Shendre, A.; Storniolo, A.M.; He, C.; Tanaka, H. A plasma telomeric cell-free DNA level in unaffected women with BRCA1 or/and BRCA2 mutations: A pilot study. Oncotarget 2018, 9, 4214–4222.
- Shi, Y.; Zhang, Y.; Zhang, L.; Ma, J.-L.; Zhou, T.; Li, Z.-X.; Liu, W.-D.; Li, W.-Q.; Deng, D.-J.; You, W.-C.; et al. Telomere Length of Circulating Cell-Free DNA and Gastric Cancer in a Chinese Population at High-Risk. Front. Oncol. 2019, 9, 1434.
- Benati, M.; Montagnana, M.; Danese, E.; Mazzon, M.; Paviati, E.; Garzon, S.; Laganà, A.S.; Casarin, J.; Giudici, S.; Raffaelli, R.; et al. Aberrant Telomere Length in Circulating Cell-Free DNA as Possible Blood Biomarker with High Diagnostic Performance in Endometrial Cancer. Pathol. Oncol. Res. 2020, 26, 2281–2289.
- Thakur, S.; Tobey, A.; Daley, B.; Auh, S.; Walter, M.; Patel, D.; Nilubol, N.; Kebebew, E.; Patel, A.; Jensen, K.; et al. Limited Utility of Circulating Cell-Free DNA Integrity as a Diagnostic Tool for Differentiating between Malignant and Benign Thyroid Nodules with Indeterminate Cytology (Bethesda Category III). Front. Oncol. 2019, 9, 905.
- Zhu, F.; Ma, J.; Ru, D.; Wu, N.; Zhang, Y.; Li, H.; Liu, X.; Li, J.; Zhang, H.; Xu, Y.; et al. Plasma DNA Integrity as a Prognostic Biomarker for Colorectal Cancer Chemotherapy. J. Oncol. 2021, 2021, 5569783.
- Deligezer, U.; Eralp, Y.; Akisik, E.E.; Akisik, E.Z.; Saip, P.; Topuz, E.; Dalay, N. Size distribution of circulating cell-free DNA in sera of breast cancer patients in the course of adjuvant chemotherapy. Clin. Chem. Lab. Med. 2008, 46, 311–317.
- Lehner, J.; Stötzer, O.J.; Fersching, D.; Nagel, D.; Holdenrieder, S. Circulating plasma DNA and DNA integrity in breast cancer patients undergoing neoadjuvant chemotherapy. Clin. Chim. Acta 2013, 425, 206–211.
- Agostini, M.; Pucciarelli, S.; Enzo, M.V.; Del Bianco, P.; Briarava, M.; Bedin, C.; Maretto, I.; Friso, M.L.; Lonardi, S.; Mescoli, C.; et al. Circulating Cell-Free DNA: A Promising Marker of Pathologic Tumor Response in Rectal Cancer Patients Receiving Preoperative Chemoradiotherapy. Ann. Surg. Oncol. 2011, 18, 2461–2468.




