Homeobox (HOX) genes are master regulators of morphogenesis and cell differentiation. All such genes contain a “homeobox”, which is a DNA sequence of roughly 180 base pairs that encodes the homeodomain, which comprises three helical regions folded into a tight globular structure that binds a 5’-TAAT-3’ core motif. The homeodomain is highly conserved and is identical from flies to humans
[1] and this 60-amino-acid motif is responsible for binding to target genes to activate transcription
[2][3][4]. As transcription factors, homeodomain proteins are involved in embryonic patterning and cell differentiation, and many have been connected to human diseases
[5]. Based on the percentage of sequence similarity, homeobox genes are classified into several families, such as HOX, NKX, PAX, GBX2, and MSX
[6]. Different functional properties amongst families are determined by DNA-binding specificities, co-factors, and other mechanisms, such as additional domains or motifs found within homeoprotein families
[6].
Homeobox genes are necessary for diverse biological functions as they control the expression of a wide range of genes that play crucial roles in proliferation, differentiation, survival, cell cycle, migration, and apoptosis. Consequently, there are many target genes with a variety of known functions, and these include p53, p21, VEGF, osteopontin, IGFBP1, NCAM, keratins, and integrins
[2]. The different homeodomain families possess varied biological functions. For example, MSX genes regulate epithelial to mesenchymal transition during organogenesis
[7], whereas members of NKX, PAX, and DLX are involved in differentiation in a tissue-specific manner
[6]. Homeodomain proteins are not specific to adult differentiated cells but are also seen in adult stem cells, such as CD34+ hematopoietic stem cells
[8]. In particular, the crosstalk between embryonic- and differentiation-specific homeodomain proteins provides a link between organogenesis and cancer.
2. HOPX in Carcinogenesis
2.1. HOPX as a Tumor Suppressor Gene
There is now a wide body of evidence, which the researchers outline below, that indicates that homeodomain-only protein (HOPX) functions to suppress the carcinogenic process. The expression, methylation status, and function of HOPX in different cancer types is summarized in Table 1.
Table 1. Deregulation and functional significance of HOPX in cancer.
| Type of Cancer |
Expression Pattern and Methylation Status |
Functional Insights |
References |
| Head and Neck cancer |
|
|
[9][10][11] |
| Esophageal cancer |
|
|
[10] |
| Gastric cancer |
|
-
Regulates proliferation, apoptosis, invasion, and tumorigenesis (in vitro).
-
It has prognostic significance.
|
[12] |
| Lung Cancer |
|
-
Regulates proliferation, apoptosis, invasion, metastasis, and tumorigenesis (both in vitro and in vivo).
-
It induces senescence through the RAS/MAPK pathway.
-
HOPX synergizes with GATA6 to regulate metastasis and angiogenesis of lung cancer cells.
-
Downregulation of HOPX correlates with the differentiation status and relapses.
|
[13][14][15][16] |
| Uterine cancer |
|
|
[17][18] |
| Colon cancer |
-
Hypermethylated and downregulated.
-
Normal stromal cells showed high levels of HOPX expression, and it was low in cancer cells.
|
-
Regulates proliferation, apoptosis, angiogenesis, invasion, and tumorigenesis (both in vitro and in vivo).
-
It has prognostic significance.
|
[19][20] |
| Pancreatic cancer |
|
|
[21] |
| Breast cancer |
|
|
[18] |
| Placenta (Tropho) |
|
|
[17] |
| Glioblastoma |
|
|
[22] |
2.1.1. HOPX Expression in Normal and Malignant Tissues
HOPX was initially isolated using a PCR-based subtracted fragmentary cDNA library between normal placental villi and a choriocarcinoma cell line
[17]. HOPX expression was later detected in normal tissues of the lung, smooth muscle, brain, placenta, bladder, spleen, and kidney
[17]. Subsequently, HOPX was shown to be downregulated in a wide range of human tumor types, including lung cancer
[16], choriocarcinoma
[17], head and neck cancers
[9][11][23][24][25][26], esophageal squamous cell carcinoma
[10], glioblastoma
[22], human uterine endometrial cancers (HECs)
[18], gastric cancer
[12], colorectal cancer
[19] , and pancreatic cancer
[21], suggesting a tumor suppressor role for HOPX.
More direct evidence for a tumor suppressor role for HOPX has come from
in vivo studies, where ectopic expression of HOPX in choriocarcinoma
[17], esophageal
[10], glioblastoma
[22], HEC
[18], gastric
[12], colorectal
[20], and lung cancer
[13] cells resulted in decreased tumor growth suppression in vivo. Interestingly, loss of HOPX expression has clinical relevance, at least in lung cancer, because decreased HOPX expression in lung SCC has been linked to high-grade and advanced-stage cancers
[16]and miR-421 has been shown to target HOPX, which then stimulates the Wnt/catenin signaling pathway, promoting the development of non-small cell lung cancer
[16].
Despite its function as a tumor suppressor, HOPX gene mutations appear to be rare in cancer
[18]. The precise role of the individual splice variants is still unknown; however, all variants were detected in HNSCCs
[26], and in skin cancer
[27] and acute myeloid leukemia
[28], the level of transcripts 1, 3, and 5 were reported to be low while variants 2 and 4 were predominantly expressed.
2.1.2. HOPX Promoter Methylation
Methylation of the cytosine residues of the CpG island within the promoter region of tumor suppressor genes is one of the mechanisms that suppresses their function in cancer
[29]. HOPX promoter methylation has been detected in human uterine endometrial cancer
[18], head and neck cancer
[26], gastric cancer
[12], colorectal cancer
[19][20], pancreatic cancer
[21], and lung cancer
[13]. Methylation of the HOPX promoter can occur as an early event in carcinogenesis because it has been detected in the early stages of gastric cancer
[12]. Interestingly, HOPX has been ranked second among the top priority genes that undergo methylation in primary esophageal squamous cell carcinoma
[10]. Treatment of head and neck, esophageal, and pancreatic cancer cells with demethylating drugs such as 5-aza-2-deoxycytidine restored HOPX expression
[10][21][26]. HOPX does not harbor mutations in human uterine endometrial cancer; however, its hypermethylation demonstrates epigenetic modification as a crucial event for inactivation
[18]. Loss of HOPX expression as a result of promoter hypermethylation has clinical significance because in patients with gastric and colorectal cancer, the methylation status has been linked to a worse prognosis
[12][20] and patients with strong HOPX expression in lung cancer have a considerably longer survival
[13]. Epigenetic suppression of HOPX by promoter methylation plays a critical role in aggressive phenotypes in papillary thyroid cancer and is correlated with a poor prognosis of patients
[30].
2.1.3. HOPX Affects Tumor Cell Behavior
Numerous in vitro and in vivo studies have shown that HOPX affects a variety of cellular processes and loss of HOPX promotes the malignant phenotype of tumor cells by a variety of mechanisms, as summarized below. The possible signaling networks regulated by HOPX in cancer are depicted in Figure 1.
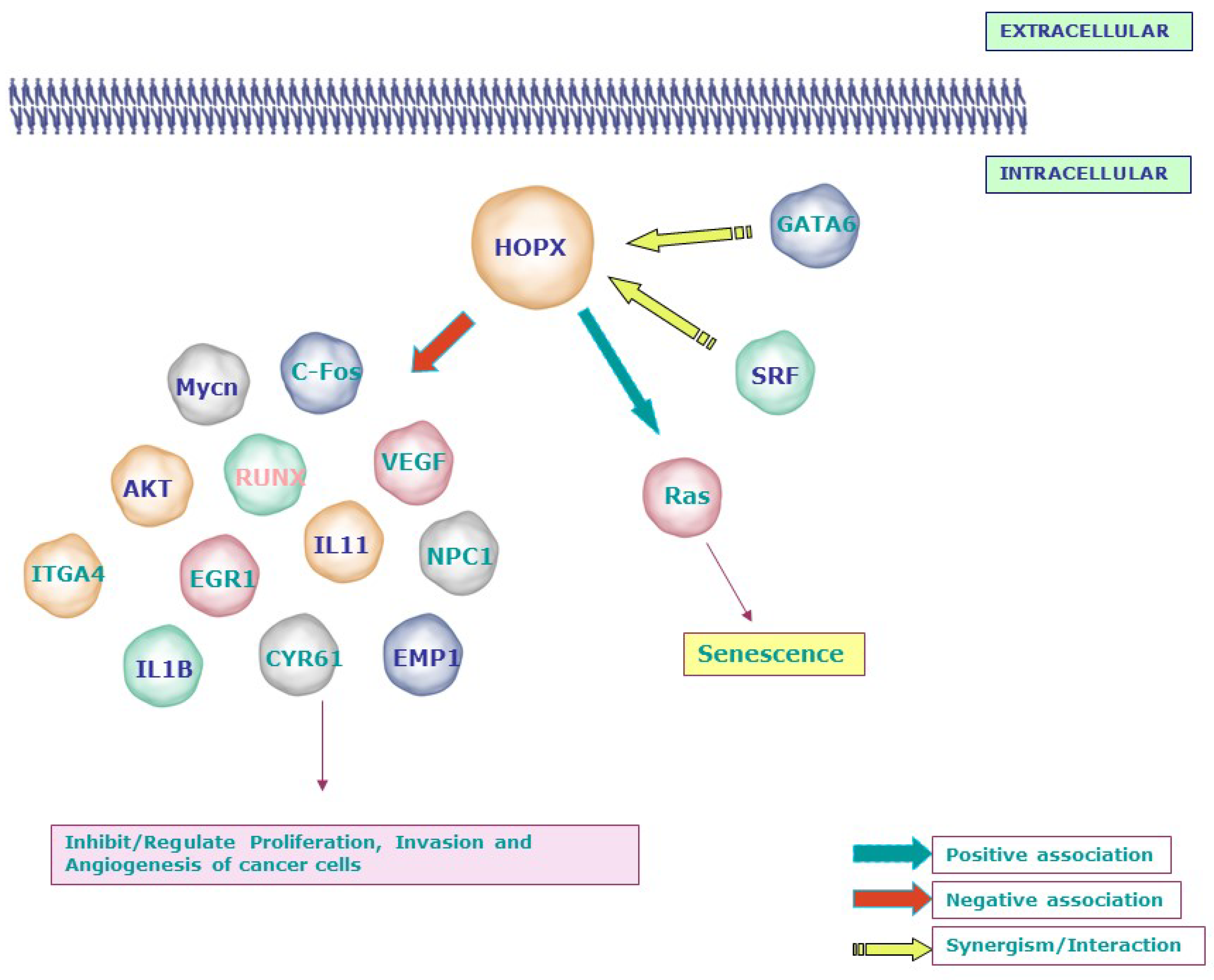
Figure 1. Signaling molecules related to the tumor suppressor function of HOPX: Loss or gain experiments and microarray analyses revealed the negative association between HOPX and molecules involved in invasion, angiogenesis, and transformation, including Mycn, c-Fos, AKT, VEGF, EGR1, IL11, IL1B, and EMP1. In line with its mechanism during development, HOPX affects the proliferation of cancer cells by synergizing with SRF and GATA6. Notably, HOPX induces senescence through the RAS-ERK-MAPK signaling cascade in lung cancer. SLC2A3—Solute Carrier Family 2 (Facilitated Glucose Transporter), Member 3; NPC1—Niemann–Pick disease, type C1; RUNX1—Runt-related transcription factor 1; CNN1/CYR61—Cysteine-rich, angiogenic inducer, 61; WTAP—Wilms tumor 1-associated protein; PRDX2—Peroxiredoxin 2; EREG—Epiregulin; ITGA4—Integrin, alpha 4; EMP1—Epithelial membrane protein 1; EPHA2—EPH receptor A2; EGR1—Early growth response; MYCN—v-myc avian myelocytomatosis viral oncogene neuroblastoma-derived homolog; IGFII—Insulin-like growth factor 2 (somatomedin A).
HOPX was found to be involved in the regulation of the cell cycle and apoptosis. By positively regulating the subG1 and GO/G1 phases, HOPX reduces DNA synthesis and promotes cell death in pancreatic cancer
[21]. Glioblastomas (GBMs) are derived from neural cancer stem cells and HOPX regulates the hippocampal stem cell niche through controlling cell survival and apoptosis, but it is downregulated in GBMs in mice; a key finding of this work is that reactivating HOPX causes apoptosis and lowers the tumorigenic ability of GBM cancer stem cells
[22]. HOPX is epigenetically silenced in breast cancer. HOPX overexpression causes apoptosis and cell cycle arrest in breast cancer cells, suggesting that it could be utilized as a therapeutic target for breast cancer patients
[31].
HOPX is activated by 17b-estradiol (E2) and inhibits cell proliferation
in vitro and
in vivo, suppressing uterine endometrial malignancies (HEC)
[18]. This occurs by influencing the activity of SRF and the expression of its targets, c-Fos and Cyclin D1. HOPX activity was restored when HEC cell lines were treated with the histone deacetylase inhibitor TSA and inhibiting HOPX also stimulated immortalized human endometrial cells to proliferate
[18]. In esophageal squamous cell carcinoma, however, HOPX had no effect on SRF activity, showing that the tumor suppressor functions of HOPX are cell type dependent
[10]. DNA microarray analysis in colorectal cancer cells showed that the re-expression of HOPX upregulates WTAP and PRDX2 while downregulating genes associated with proliferation, angiogenesis, and invasion both in vitro and in vivo
[20].
HOPX expression has been reported to be reduced at both the DNA and protein levels of oral and nasopharyngeal cancer cells, and its re-expression promoted cell proliferation and invasion and made the cells more susceptible to UV and cisplatin-induced cell death
[26].
Interestingly, a recent study showed that the HOPX knockdown in squamous cell carcinoma (SCC) cells reduces their proliferative and invasive capabilities through the acceleration of apoptosis. The methylation status of two distinct HOPX promoters, promoter 1 and 2, causes the HOPX transcript expression to differ in normal keratinocytes and SCC cells. The activation of the second promoter of HOPX may contribute to the pathogenesis of SCC
[27].
2.1.4. HOPX and Senescence
There are a limited number of reports suggesting a role for HOPX in cellular senescence. For example, HOPX transfection of lung cancer cells resulted in enlarged and flattened morphologies, with positive SA—Gal staining indicating senescence, and stimulated the expression of proteins associated with aging, such as SAHFs and p-H2AX. When these cells were treated with senescence-inducing drugs such as Adriamycin (ADR) and H
2O
2, senescence was caused in HOPX-transfected cells but not in mock transfectants
[13]. In H2228 lung adenocarcinoma cells, however, HOPX did not cause senescence, showing that it is involved in oxidative stress and DNA damage-induced cellular senescence but does not cause senescence on its own
[13].
It was reported that re-expressing HOPX in immortalized bronchial epithelial cells promotes senescence. The RAS-ERK-MAPK signaling cascade has been identified to be involved in HOPX-induced senescence in lung cancer cells. A significant reduction of AKT/MDM2 accompanied by an upregulation of P53/ P21 was observed in HOPX-transfected lung cancer and normal bronchial epithelial cells. This research revealed HOPX as a regulator of the expression of senescence mediators
[13]. Furthermore, the HOPX gene was shown to be overexpressed in keratinocytes with shortened telomeres, which elicits senescence
[32].
2.1.5. HOPX in Cancer Cell Metastasis
A critical molecular event in the development of lung cancer is the downregulation of HOPX and overexpression of Mycn
[33], and re-expression of HOPX suppresses lung cancer cell proliferation migration and invasion
[13]. Furthermore, the alveolar lineage transcription factors, GATA6 and HOPX, were reported to be essential in airway epithelial specification and lung adenocarcinoma subtype metastasis inhibition
[14].
In a chicken metastatic cell line derived from v-src-induced sarcoma and its non-metastatic subclone, the role of HOPX in metastasis was investigated. In v-src-transformed cells, knocking down HOPX reduced metastatic activity by inactivating metastatic-associated genes such as ITGA4
[34]. The expression of EMT-related molecules, such as ZEB1/2, TWIST1, E-cadherin, and vimentin, however, was unaffected by GATA6 and HOPX knockdown. It also had no effect on Wnt or focal adhesion kinase (FAK), although the expression of src and integrin 5, two mediators of metastasis, was upregulated.
HOPX hypermethylation promotes metastasis in nasopharyngeal cancer by increasing SNAIL transcription, which facilitates epithelial to mesenchymal transition
[35]. Additional metastasis-related molecules (IL1B, IL11, and EREG) and angiogenesis and ECM remodeling genes (VEGFA and PLAU) were upregulated when HOPX was knocked down
[14].
Bioinformatic analysis using STRING software predicts the possible interacting partners of HOPX in mice and humans as indicated in
Figure 2[36].
Figure 2. Possible interaction partners of HOPX in mice and humans as predicted using the STRING database. Apart from SRF, HDAC2, and Epc1, HOPX could interact with proteins such as Apc, Nts, Ran, and Rasl2-9 in mice and COL14A1, SH3GL1, MRPL49, and HRASLS5 in humans. Mouse: Nkx2-1—NK2 homeobox 1; Ran—RAN, member RAS oncogene family; Naa50—N(alpha)-acetyltransferase 50, NatE catalytic subunit Gene; Nts—neurotensin; Srf—serum response factor; Sh3gl1—SH3-domain GRB2-like 1; Gpr182—G protein-coupled receptor 182; Rasl2-9—RAS-like, family 2, locus 9; Hopx—Homeodomain-only protein; Omp—olfactory marker protein; Apc—adenomatosis polyposis coli. Human: SRI—sorcin; HDAC2—histone deacetylase 2; COL14A1—collagen, type XIV, alpha 1; HOPX—Homeodomain-only protein; SH3GL1—SH3-domain GRB2-like 1; SRF—serum response factor (c-fos serum response element-binding transcription factor); AVPR2—arginine vasopressin receptor 2; MRPL49—mitochondrial ribosomal protein L49; MCL1—myeloid cell leukemia sequence 1 (BCL2-related); HRASLS5—HRAS-like suppressor family, member 5; AATF—apoptosis antagonizing transcription factor.
2.2. Evidence for an Oncogenic Function for HOPX
The expression level of HOPX is decreased in most cancer types and it functions as a tumor suppressor. However, HOPX expression has been reported to be elevated in a small number of esophageal carcinoma cell lines (TE2, TE5, KYSE410, and KYSE520), lung adenocarcinoma, and pancreatic cancer Langerhans islet cells
[10][13][21]. Strong HOPX expression was associated with specific clinical and biochemical characteristics in de novo acute myeloid leukemia and predicted a poor prognosis
[28]. HOPX is one of a group of genes, including deoxynucleotidyl transferase (DNTT) and B cell liners (BLNK), that are upregulated in patients with RUNX-1 mutant cytogenetically normal acute leukemia
[37]. Interestingly, HOPX expression is increased in HPV 16- and 18-infected stage IB-IIA cervical cancer cells
[38]. The expression of HOPX was also noticed in the tumor microenvironment; in particular, HOPX expression was higher in normal stromal cells than in stromal cells from colorectal malignant tissue
[19][20]. There is increasing evidence that HOPX is a tumor suppressor gene in numerous cancer types. However, the function of HOPX in cancer could well be context dependent and more studies are needed to confirm a possible oncogenic role for HOPX in certain circumstances.