Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Qingzhong Wang | -- | 2604 | 2022-06-07 13:26:57 | | | |
| 2 | Conner Chen | -20 word(s) | 2584 | 2022-06-08 02:22:58 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Wang, Q. m6A RNA Methylation Mechanism. Encyclopedia. Available online: https://encyclopedia.pub/entry/23782 (accessed on 03 March 2026).
Wang Q. m6A RNA Methylation Mechanism. Encyclopedia. Available at: https://encyclopedia.pub/entry/23782. Accessed March 03, 2026.
Wang, Qingzhong. " m6A RNA Methylation Mechanism" Encyclopedia, https://encyclopedia.pub/entry/23782 (accessed March 03, 2026).
Wang, Q. (2022, June 07). m6A RNA Methylation Mechanism. In Encyclopedia. https://encyclopedia.pub/entry/23782
Wang, Qingzhong. " m6A RNA Methylation Mechanism." Encyclopedia. Web. 07 June, 2022.
Copy Citation
Epitranscriptomic modifications can affect every aspect of RNA biology, including stability, transport, splicing, and translation, participate in global intracellular mRNA metabolism, and regulate gene expression and a variety of biological processes. N6-methyladenosine (m6A) as the most prevalent modification contributes to normal embryonic brain development and memory formation.
m6A-modifying enzymes
Epitranscrptome
Neuropsychiatric disorders
Drug discovery based on m6A related enzymes
1. Introduction
Epigenetic modification refers to the reversible and heritable changes in gene function under the condition of an unchanged gene sequence and mainly includes DNA methylation and histone modification [1][2][3][4][5]. Similar to DNA epigenetics, RNA epigenetics also involves RNA chemical modifications that regulate gene expression by affecting RNA stability and translation [6]. To date, more than 150 types of RNA modifications have been identified, such as N1-methyladenosine [7], N7-methylguanosine [8], 5-methylcytosine [9], N6,2-dimethyladenosine glycosides [10][11], etc. These RNA modifications are widely distributed in various types of RNA, including messenger RNA (mRNA), transfer RNA (tRNA), ribosomal RNA (rRNA), small non-coding RNA, and long non-coding RNA (lncRNA) [12]. Among them, m6A modification is the most common chemical modification on mRNA discovered in recent years [13]. Studies have shown that m6A modification is involved in the metabolic activity of intracellular mRNA, regulates gene expression, and controls various biological processes such as RNA stability and mRNA translation [14]. Nevertheless, the detailed mechanisms of m6A modification have not been fully elucidated.
It has been reported that m6A modification is highly enriched in the brain and plays an important role in central nervous system development and neurodegenerative diseases [15]. The m6A modification is involved in the biological process of the central nervous system by regulating neural-related mRNA expression. When the activity or expression level of m6A-modifying enzymes in the brain is altered, the m6A modification level of related mRNAs is disrupted, leading to blockage of nuclear export, splicing, translation, and other processes, which in turn results in the occurrence of central nervous system diseases [16].
2. m6A RNA Methylation Mechanism
Desrosiers et al., 1975 first proposed a new RNA epigenetic modification, N6-methyladenosine (m6A), when analyzing the polyadenylic acid structure in a tumor cell, and found that approximately 80% of mRNA epitranscriptomic changes are m6A methylation modifications [17]. Since then, the presence of m6A has been detected in the RNAs of various eukaryotes and viruses such as yeast and Arabidopsis [18][19][20]. Recent studies have revealed that there are more than 7000 m6A-modified mRNAs in mammalian cells, and m6A also exists in ribosomal RNAs, transfer RNAs, small nucleolar RNAs, miRNAs, and long non-coding RNAs [12]. A comprehensive analysis of mRNA methylation has shown that the distribution of m6A in protein-coding regions, untranslated regions, and introns was 50.9%, 41.9%, and 2.0%, respectively. In the protein-coding region, m6A is mainly enriched near the stop codon, while in the untranslated region, m6A is mainly enriched at the 3’ end [11]. Analysis of the results of m6A high-throughput sequencing shows that m6A modification commonly occurs on the sequence of adenine of RRACH motif (R represents A or G, H represents U, A, or C) [10]. In mammals, the content of m6A modification varies in different tissues and is especially enriched in the liver, kidney, and brain [21]. In recent years, with the advancement of m6A modification detection methods, the mechanism of m6A modification has been gradually uncovered. The dynamic regulation mechanism of m6A modification mainly relies on three enzymes, namely methyltransferase complexes (“Writers”), demethylases (“Erasers”), and methyl readers (“Readers”) (Figure 1). Writers use S-adenosylmethionine (SAM) as a methyl donor to catalyze the formation of a methyl group at the sixth N element of adenine in RNA. At the same time, this modification can also be removed by an eraser to achieve dynamic adjustment. Readers are responsible for recognizing modification signals, which in turn affect the processes of RNA export, splicing, translation, and degradation.
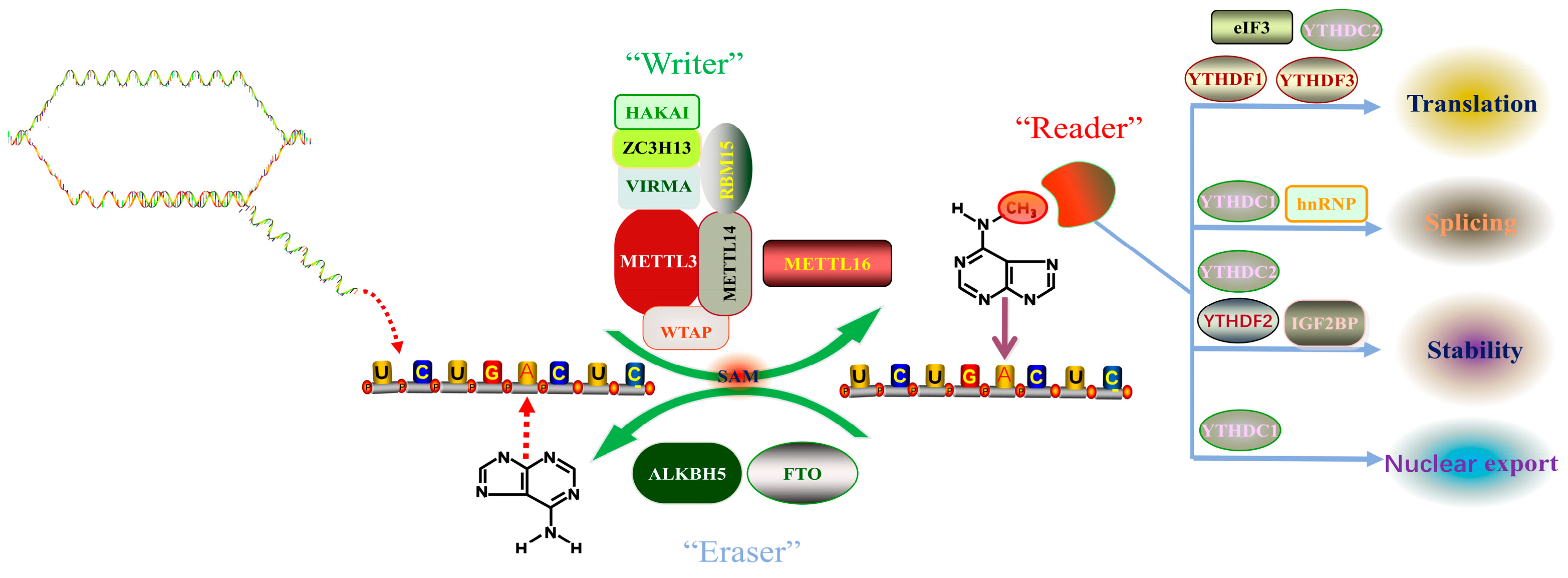
Figure 1. Dynamic m6A modification of RNA. Abbreviations: METTL3 (Methyltransferase-like protein 3); METTL14 (Methyltransferase-like protein 14); WTAP (Wt1 Associated Protein); KIAA1429 (Vir Like M6A Methyltransferase Associated, VIRMA); RBM15 (RNA Binding Motif Protein 15); HAKAI (Cbl Proto-Oncogene Like 1); ZC3H13 (Zinc Finger Ccch-Type Containing 13); METTL16 (Methyltransferase-like protein 16); FTO (Fat Mass and Obesity Associated Protein); ALKBH5 (Alkb Homolog 5, RNA Demethylase); eIF3 (eukaryotic initiation factor 3); hnRNP (heterogeneous nuclear ribonucleoprotein); YTHDF2 (Yth N6-Methyladenosine RNA Binding Protein 2); YTHDF1 (Yth N6-Methyladenosine RNA Binding Protein 1); YTHDF3 (Yth N6-Methyladenosine RNA Binding Protein 3); YTHDC1 (Yth Domain Containing 1); YTHDC2 (Yth Domain Containing 2).
2.1. m6A Methyltransferase (Writers)
The m6A-modified methyltransferase complex is mainly composed of METTL3, METTL14, WTAP, and KIAA1429 and also includes ZFP217 (Zinc Finger Protein 217), RBM15, RBM15B (RNA Binding Motif Protein 15B), HAKAI, ZC3H13, and other components [22][23]. Among them, METTL3 is the core component of methyltransferase that plays a catalytic role, and METTL14 is responsible for RNA recruitment, forms a heterodimer with METTL3, and jointly promotes the production of m6A modification. WTAP is responsible for stabilizing the complex, while RBM15 and RBM15B assist METTL3 in binding to WTAP so that they can precisely localize the target site and participate in the production of the m6A modification.
The m6A methyltransferase enzyme complex was first isolated with artificially synthesized RNA fragments from Hela cells, named MT-A (200 kDa) and MT-B (800 kDa), respectively [24][25]. Subsequently, MT-A1 and MT-A2 were successfully separated from MT-A. MT-A1 has a smaller molecular weight, only 30 kDa, and no methylase activity was found; MT-A2 as a multimer has a molecular weight of 200 kDa. The MT-A protein includes a 70 kDa protein with an S-adenosylmethionine binding site (MT-A70), which has the function of catalyzing m6A modification. MT-B has the largest molecular weight (875 kDa) in the enzyme complex, and has the properties of an RNA-binding protein, but its catalytic activity is low, and in the absence of MT-A1 and MT-A2, MT-B methylase activity disappeared [25]. The MT-A70 subunit contains two motifs, the S-adenosylmethionine (SAM) binding site and the DPPW motif (Asp-Pro-Pro-Trp) functional domain with catalytic function [25][26]. Immunofluorescence showed that METTL3 was mainly located in the nuclear speckles region in the nucleus, and this region was highly enriched for mRNA splicing factors, which indicated the potential role of METTL3 in the regulation of RNA metabolism [25].
METTL14 is an m6A-catalyzed methyltransferase discovered by homology analysis of the MT-A70 family [27]. Studies have shown that knockdown of METTL14 can reduce the extent of m6A modification in Hela cells and 293T cells, suggesting that METTL14 can catalyze the methylation reaction of m6A [28]; METTL3 and METTL14 can form a stable heterodimeric core complex with equal amounts, which functions as a methylase, and the complex of METTL3 and METTL14 has a strong affinity for the conserved sequence GGACU modified by m6A. METTL14 alone has higher m6A methylase activity than METTL3 alone, and when these two proteins combine into a heterodimer, the catalytic activity is significantly increased, and both are colocalized in the nuclear plaque region of the nucleus, playing a synergistic role in maintaining the stability of each other’s proteins. The downregulation of METTL3 or METTL4 leads to increased expression of certain target genes, suggesting that m6A may reduce mRNA stability [28]. Further investigation of the structure of the METTL3-METTL14 complex revealed that METTL3 has a catalytic subunit that serves mainly as the core of m6A’s catalytic activity, while METTL14 is mainly responsible for RNA recognition and binding while stabilizing the structure of the complex [22][29].
WTAP is the third identified m6A-modifying enzyme component. Originally, WTAP functions as a splicing factor for WT1 (Wilms tumor 1) protein and regulates cell cycle and embryonic development [30]. In Arabidopsis, the homologous protein mRNA adenosine methylase (MTA) of METTL3 was first found to interact with FIP37 (FKBP12 interacting protein 37) in vitro and in vivo, and FIP37 is the homologous protein of WTAP [31]. Meanwhile, Liu et al. (2014) found that WTAP can bind to the heterodimeric METTL3-METTL14 core complex and affect its catalytic activity of m6A modification [28]. Ping et al., 2014 found that WTAP interacts with the METTL3-METTLl4 complex, which may contribute to its localization in the pre-mRNA enriched nuclear plaque region and is essential for the catalytic activity of m6A. Under the absence of WTAP, the RNA-binding ability of METTL3 decreased significantly, suggesting that WTAP may regulate the recruitment of the m6A methyltransferase complex to mRNA targets [23]. METTL3 and METTL14 mainly bind the GGAC motif, WTAP mainly binds the GACU motif, and the three proteins share approximately 36% of the common binding motif [28].
RBM15 is another component of the m6A methyltransferase complex. Proteomic analysis revealed that RBM15, RBM15B, and WTAP may have a synergistic effect on m6A methyltransferase. The knockout of WTAP reduces the interaction between METTL3 and RBM15. Down-regulated expression of RBM15 and RBM15B significantly decreased m6A modification and reduced XIST-mediated downregulation [32].
2.2. m6A Demethylases (Erasers)
Similar to DNA and histone methylation, RNA has m6A demethylases that remove m6A-modified methyl groups. By 2011, FTO and ALKBH5 have been confirmed to be m6A-modified demethylating enzymes [33]. FTO was first discovered while studying fused toe mutations in mice [34], and subsequent studies found that the absence of FTO in mice leads to increased energy expenditure and systemic sympathetic activation. This leads to stunted growth and a marked reduction in adipose tissue and lean body mass [35]. In humans, mutations in FTO lead to a significant increase in body mass index, obesity, and a predisposition to diabetes [36]. Gerken et al., 2007 found that FTO belongs to the family of Fe(II) and 2-OG-dependent dioxygenases that catalyze the demethylation of single-stranded DNA [37]. In 2011, Jia et al., 2011 found that FTO can effectively demethylate m6A and that the knockout of FTO leads to an increase in the amount of m6A in mRNA, demonstrating for the first time the demethylase effect of FTO [33]; Xu et al., 2014 found that the expression of FTO significantly negatively correlated with the degree of m6A modification during adipogenesis. Downregulation of FTO can increase the level of m6A modification of the alternative splicing regulator SRSF2 (Serine and Arginine Rich Splicing Factor 2) and improve its RNA-binding ability. Additionally, FTO affects the exon splicing of the adipogenic regulator RUNX1T1 (Runx1 Partner Transcriptional Co-Repressor 1) by regulating the level of m6A near the splice site, thereby regulating adipose progenitor cell differentiation [38].
ALKBH5, the second m6A demethylase discovered in 2013, reverses the m6A modification of mRNA in vitro and in vivo. In HeLa cells, the downregulation of ALKBH5 increased m6A content in total mRNA by 9% and the overexpression of ALKBH5 decreased m6A content in total mRNA by 29% [39]. In addition, ALKBH5 can affect mRNA processing, nuclear export, and RNA metabolism through demethylase activity, and the level of mRNA m6A in mice with ALKBH5 gene deficiency is significantly increased, leading to apoptosis of spermatocytes in mid-meiosis and significantly weakening reproductive ability [39].
2.3. m6A Binding Proteins (Readers)
The main function of reader proteins is to recognize bases undergoing m6A modification and thus regulate the biological function of RNA. The identified m6A-binding proteins mainly include YTH domain-containing RNA-binding proteins, hnRNP, eIF3, IGF2BPs (insulin-like growth factor 2 mRNA-binding proteins), etc., which can specifically recognize and bind to m6A-modified mRNA and then play regulatory roles in mRNA stability, export, translation, splicing, and degradation [40][41][42][43][44].
YTHDF2 is the first m6A-binding protein to be discovered. Studies have shown that m6A can regulate the stability, processing, or translation of m6A-modified RNAs mainly by recruiting specific reader proteins or altering the RNA structure. YT521-B homology domain family (YTHDF) proteins are cytoplasmic recognizers of m6A that can specifically recognize and bind m6A-modified RRCH sequences and affect the stability and translation efficiency of RNAs containing m6A modifications; The direct interaction between the N-terminal region of CNOT1 and the SH region of the CNOT1 (Ccr4-Not Transcription Complex Subunit 1) subunit regulate target mRNA deadenylation and degradation [45][46].
YTHDF1 is another m6A-recognizing protein that was later identified. Investigating other functional roles of m6A modification, Wang et al., 2015 discovered another m6A-binding protein, YTHDF1, that can selectively recognize m6A-modified mRNA, promote its loading on ribosomes, enhance binding to translation initiation factor eIF3 and translation initiation, and affect gene expression by influencing mRNA translation and degradation [47].
It was also confirmed that YTHDF3 is an m6A-binding protein that promotes the translation of m6A-modified mRNA and may play a role in the initiation phase of translation via ribosomal proteins. The translation efficiency of the common targets of YTHDF1 and YTHDF3 was significantly enhanced, suggesting that the two may play a synergistic role in the translation process to produce their regulatory functions and that the effect of YTHDF3 on its own specific targets may be related to other RNA processing [48][49].
YTHDC1 is mainly located in the nucleus and can form YT bodies around the active transcription site and RNA processing structures. It was later found to bind specifically to m6A-modified RNA and regulate mRNA splicing; YTHDC1 and YTHDF2 share 21% of the m6A binding site. YTHDC1 has a stronger binding force to m6A modification and can easily bind to the GG (m6A) C sequence, and most of its binding sites are located around the stop codon [42]. Xiao et al., 2016 found that YTHDC1 can inhibit the binding of splicing factor SRSF10 (Serine and arginine rich splicing factor 10) to target mRNA by promoting the binding of SRSF3 (Serine and arginine rich splicing factor 3) mRNA, increasing its exons, and then regulating mRNA splicing [50].
The cytoplasmic m6A-binding protein YTHDC2 is critical for regulating m6A transcripts to ensure successful meiotic gene expression programs in mammalian reproduction [51]. Hsu et al., 2017 and Jain et al., 2018 found that YTHDC2 can specifically recognize m6A modification sites, promote translation of its target, and reduce its mRNA abundance [52][53].
The IGF2BPs family includes three RNA-binding proteins (RBPs), IGF2BP1 (Insulin Like Growth Factor 2 mRNA Binding Protein 1), IGF2BP2 (Insulin Like Growth Factor 2 mRNA Binding Protein 2), and IGF2BP3 (Insulin Like Growth Factor 2 mRNA Binding Protein 3) [54]. Huang et al. (2018) reported that the IGF2BPs family can recognize the modification site of m6A, function as an m6A-binding protein to bind to m6A-modified mRNA transcripts, and increase the stability of target mRNAs, such as MYC (Myc Proto-Oncogene, Bhlh Transcription Factor), thereby regulating gene expression [44].
The hnRNP family also contains three RNA-binding proteins, HNRNPC (Heterogeneous Nuclear Ribonucleoprotein C), HNRNPA2B1 (Heterogeneous nuclear ribonucleoprotein A2/B1), and HNRNPG (RNA binding motif protein X-linked). Among them, HNRNPC is an RNA-binding protein in the nucleus that is mainly responsible for processing pre-mRNA, m6A-modified mRNA, and lncRNA, and can alter their local structure to then regulate gene expression by regulating the alternative splicing of exons [55]. HNRNPA2B1, a member of the hnRNP family of RNA-binding proteins, has the ability to bind m6A-modified RNA substrates, and is involved in the regulation of biological processes such as miRNA precursor processing and alternative splicing of mRNA [56]. HNRNPA2B1 specifically recognizes the m6A site sequence RGACH (R = G/A, H = A/C/U), and then promotes the interaction with the pri-miRNA microprocessor complex protein DGCR8 (DGCR8 microprocessor complex subunit), promotes pri-miRNA maturation, and then regulates microRNA expression [56][57][58]. Subsequently, Wu et al., 2018 reported the crystal structures of HNRNPA2B1 in a complex with various RNA targets and elucidated the molecular basis of specific and multivariant recognitions of RNA substrates. The results of biochemistry and bioinformatics analysis suggested that m6A switches may be responsible for enhancing the accessibility of HNRNPA2B1 binding to specific binding sites [59]. Moreover, HNRNPG has a low-complexity AGG (Arg-Gly-Gly) structural domain that binds to RNAs with m6A modifications and regulates their stability and expression [60].
References
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396.
- Ehrlich, M.; Gama-Sosa, M.A.; Carreira, L.H.; Ljungdahl, L.G.; Kuo, K.C.; Gehrke, C.W. DNA methylation in thermophilic bacteria: N 4-methylcytosine, 5-methylcytosine, and N 5 methyladenine. Nucleic Acids Res. 1985, 13, 1399–1412.
- Morgan, M.A.; Shilatifard, A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 2020, 52, 1271–1281.
- Bird, A. Methylation talk between histones and DNA. Science 2001, 294, 2113–2115.
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45.
- Rosselló-Tortella, M.; Ferrer, G.; Esteller, M. Epitranscriptomics in hematopoiesis and hematologic malignancies. Blood Cancer Discov. 2020, 1, 26.
- Zhang, C.; Jia, G. Reversible RNA Modification N1-methyladenosine (m1A) in mRNA and tRNA. Genom. Proteom. Bioinform. 2018, 16, 155–161.
- Zhang, L.S.; Liu, C.; Ma, H.; Dai, Q.; Sun, H.L.; Luo, G.; Zhang, Z.; Zhang, L.; Hu, L.; Dong, X. Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol. Cell 2019, 74, 1304–1316.e8.
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033.
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206.
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646.
- Lee, M.; Kim, B.; Kim, V.N. Emerging roles of RNA modification: m6A and U-tail. Cell 2014, 158, 980–987.
- Yue, Y.; Liu, J.; He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015, 29, 1343–1355.
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306.
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m6A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020, 21, 36–51.
- Pan, T.; Wu, F.; Li, L.; Wu, S.; Zhou, F.; Zhang, P.; Sun, C.; Xia, L. The role m6A RNA methylation is CNS development and glioma pathogenesis. Mol. Brain 2021, 14, 119.
- Desrosiers, R.C.; Friderici, K.H.; Rottman, F.M. Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5’terminus. Biochemistry 1975, 14, 4367–4374.
- Dubin, D.T.; Taylor, R.H. The methylation state of poly A-containing-messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975, 2, 1653–1668.
- Krug, R.M.; Morgan, M.A.; Shatkin, A.J. Influenza viral mRNA contains internal N6-methyladenosine and 5’-terminal 7-methylguanosine in cap structures. J. Virol. 1976, 20, 45–53.
- Colonno, R.J.; Stone, H.O. Methylation of messenger RNA of Newcastle disease virus in vitro by a virion-associated enzyme. Proc. Natl. Acad. Sci. USA 1975, 72, 2611–2615.
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326.
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 2016, 534, 575–578.
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189.
- Tuck, M.T. Partial purification of a 6-methyladenine mRNA methyltransferase which modifies internal adenine residues. Biochem. J. 1992, 288, 233–240.
- Bokar, J.A.; Rath-Shambaugh, M.E.; Ludwiczak, R.; Narayan, P.; Rottman, F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994, 269, 17697–17704.
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247.
- Bujnicki, J.M.; Feder, M.; Radlinska, M.; Blumenthal, R.M. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA: m6A methyltransferase. J. Mol. Evol. 2002, 55, 431–444.
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X. A METTL3–METTL14 complex mediates mammalian nuclear RNA N 6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95.
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 2016, 63, 306–317.
- Little, N.A.; Hastie, N.D.; Davies, R.C. Identification of WTAP, a novel Wilms’ tumour 1-associating protein. Hum. Mol. Genet. 2000, 9, 2231–2239.
- Zhong, S.; Li, H.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M.; Fray, R.G. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288.
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373.
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G. N 6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887.
- Peters, T.; Ausmeier, K.; Rüther, U. Cloning of Fatso (Fto), a novel gene deleted by the Fused toes (Ft) mouse mutation. Mamm. Genome 1999, 10, 983–986.
- Fischer, J.; Koch, L.; Emmerling, C.; Vierkotten, J.; Peters, T.; Brüning, J.C.; Rüther, U. Inactivation of the Fto gene protects from obesity. Nature 2009, 458, 894–898.
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Elliott, K.S.; Lango, H.; Rayner, N.W. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007, 316, 889–894.
- Gerken, T.; Girard, C.A.; Tung, Y.C.L.; Webby, C.J.; Saudek, V.; Hewitson, K.S.; Yeo, G.S.H.; McDonough, M.A.; Cunliffe, S.; McNeill, L.A. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 2007, 318, 1469–1472.
- Zhao, X.; Yang, Y.; Sun, B.F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.J.; Ping, X.L.; Chen, Y.S.; Wang, W.J. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419.
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29.
- Haussmann, I.U.; Bodi, Z.; Sanchez-Moran, E.; Mongan, N.P.; Archer, N.; Fray, R.G.; Soller, M. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 2016, 540, 301–304.
- Wang, X.; He, C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014, 11, 669–672.
- Xu, C.; Wang, X.; Liu, K.; Roundtree, I.A.; Tempel, W.; Li, Y.; Lu, Z.; He, C.; Min, J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014, 10, 927–929.
- Zhu, T.; Roundtree, I.A.; Wang, P.; Wang, X.; Wang, L.; Sun, C.; Tian, Y.; Li, J.; He, C.; Xu, Y. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014, 24, 1493–1496.
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295.
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120.
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 2016, 7, 12626.
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388–1399.
- Li, A.; Chen, Y.S.; Ping, X.L.; Yang, X.; Xiao, W.; Yang, Y.; Sun, H.Y.; Zhu, Q.; Baidya, P.; Wang, X. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017, 27, 444–447.
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328.
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 2016, 61, 507–519.
- Wojtas, M.N.; Pandey, R.R.; Mendel, M.; Homolka, D.; Sachidanandam, R.; Pillai, R.S. Regulation of m6A transcripts by the 3′→ 5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell 2017, 68, 374–387.e12.
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127.
- Jain, D.; Puno, M.R.; Meydan, C.; Lailler, N.; Mason, C.E.; Lima, C.D.; Anderson, K.V.; Keeney, S. ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. eLife 2018, 7, e30919.
- Bell, J.L.; Wächter, K.; Mühleck, B.; Pazaitis, N.; Köhn, M.; Lederer, M.; Hüttelmaier, S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell. Mol. Life Sci. 2013, 70, 2657–2675.
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 2015, 518, 560–564.
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 2015, 162, 1299–1308.
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008, 582, 1977–1986.
- Haley, B.; Paunesku, T.; Protić, M.; Woloschak, G.E. Response of heterogeneous ribonuclear proteins (hnRNP) to ionising radiation and their involvement in DNA damage repair. Int. J. Radiat. Biol. 2009, 85, 643–655.
- Wu, B.; Su, S.; Patil, D.P.; Liu, H.; Gan, J.; Jaffrey, S.R.; Ma, J. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018, 9, 420.
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.0K
Revisions:
2 times
(View History)
Update Date:
08 Jun 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





