
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Gerardo Perozziello | -- | 633 | 2022-06-06 10:40:29 | | | |
| 2 | Conner Chen | + 2676 word(s) | 3309 | 2022-06-27 08:39:12 | | |
Video Upload Options
Traditional cell cultures are performed in two-dimensional (2D) systems such as Petri dishes, multiwell plates or flasks. However, they cannot realistically mimic the morphophysiological complexity of the original three-dimensional (3D) in vivo environment from which the cells of specific lines originate. Without opposing animal experimentation but promoting its responsible application, the development of alternative cell culture systems tries to ensure compliance with the 3R principles. Reduction (reduction in the animals used for in vivo tests), Refinement (experimental design optimization to limit stress and affliction to laboratory animals) and Replacement (total or partial replacement of animal testing with alternative valid methods) are increasingly desired and strongly recommended as fundamental ethical aspects in the use of animals in scientific experiments.
1. Introduction
2. Physiological Exchange of Substances
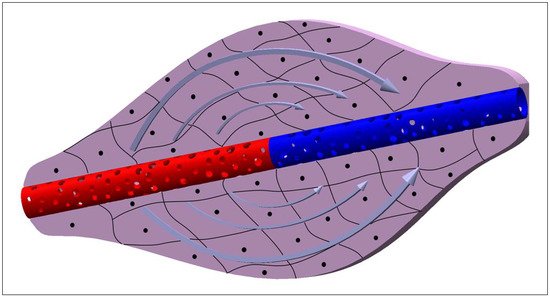
3. Theory behind the Molecule Transport Mechanisms
- ΔC=(C2−C1) is the concentration gradient of a generic molecule between the external and internal part of the capillary membrane;
- P is the permeability coefficient and can be calculated as:
- Δx is the capillary membrane thickness;
- α is the partition coefficient and can be calculated as:
- N is the number of pores;
- A is the capillary surface;
- R is the pore radius;
- n is the pore density;
- DM is the membrane diffusion coefficient and can be calculated as:
-
ϵ is the hindrance coefficient and it depends on the particle and membrane pore dimension and the trajectory of the particle within the pore and can be calculated as:
- ϵ2 is a coefficient that depends on the trajectory of the particle inside the pore;
- r is the particle radius (it is an approximation which considers the molecules passing the pore to have a spherical shape);
- D is the diffusion coefficient which can be calculated as:
- k is the Boltzman constant;
- T is the temperature;
- η is the blood viscosity;
- Δp=(p2−p1) is the hydraulic pressure gradient across the capillary membrane;
- Δπ=(π2−π1) is the osmotic pressure gradient across the capillary membrane;
- Lp is the filtration coefficient and can be calculated as:
4. Cell Microenvironment: Static and 3D Cell Screening
| Microfluidic Platform Type | Application | Cell Lines | References |
|---|---|---|---|
| Resin 3D-printed system (VeroClear, MED610 resins) |
Cell Culture, LC-MS/MS single cell analysis | BPAECs (Bovine Pulmonary Artery Endothelial Cells), MDCK (Madin-Darby Canine Kidney) | [52] |
| Microwell-based PDMS-membrane-PDMS sandwich multilayer chips | Spheroid formation, OoC | C3A (liver) | [53] |
| Two-stage temperature-controlling system used to generate decellularized extracellular matrix (dECM) hydrogel microspheres | dECM hydrogels microsphere formation, cell culture | Schwann cells (nervous tissue), PC12 (adrenal gland) | [54] |
| Injection-molded Polystyrene array |
OoC, angiogenesis | HUVEC (Human Umbilical Vein Endothelial Cells), fibroblasts | [55] |
| PDMS-gut-on-a-chip device either with a straight channel or a non-linear convoluted channel, transwell-embedded hybrid chip | OoC | Caco-2 (colon) | [56] |
| Cyclo-olefin-polymer (COP) transparent bioreactor |
On-chip platelet production | imMKCLs (immortalized MegaKaryocyte progenitor Cell Lines) | [57] |
| PDMS soft lithography replicas of superficial channels 3D-printed in different resins (Clear, Model, Tough, Amber, Dental resins) | OoC | HUVEC (Human Umbilical Vein Endothelial Cells), fibroblasts | [58] |
| PDMS bone-mimicking extracellular matrix composite device | Angiogenesis, OoC | SW620 (colon), MKN74 (stomach) | [59] |
| Single-chamber commercial microfluidic device |
OoC, disease model, drug screening | Primary human hepatocytes, EA.hy926 (human endothelial), U937 (pleural effusion), LX-2 (hepatic stellate cell) | [60] |
| Collagen scaffold | OoC | Caco-2 (colon) | [61] |
| Cellulose-based device | Chemotaxis, invasion assay | A549 (lung) | [62] |
| Polymerized High Internal Phase Emulsion (polyHIPE) system | OoC | hES-MPs (human Embrionic Stem cell-derived Mesenchymal Progenitor cells) | [17] |
| OrganoPlate LiverTox™ | Drug screening, OoC | Induced pluripotent stem cell (iPSC)-derived hepatocytes (iHep), endothelial cells, THP-1 monoblast (peripheral blood) | [63] |
| Injection-molded Polystyrene array |
Drug screening | HeLa (uterus, cervix), NK-92 (peripheral blood) | [64] |
| Resin 3D-printed system (VeroClear) |
Spheroid formation | OSCC (Oral Squamous Cell Carcinoma), HepG2 (liver) | [65] |
| 3D-printed device | Circulating Tumour Cells (CTCs) isolation | MCF-7 (breast), SW480 (colon), PC3 (prostate), 293T (kidney) | [66] |
| PDMS-based device | Spheroid formation, disease model, drug screening, OoC | Rat primary hepatocytes, HSCs (Hepatic Stellate Cells) | [67] |
| PDMS-glass chip and Polycarbonate cover-plates |
Four OoC | EpiIntestinal™, HepaRG (liver), HHStec (Human primary Hepatic Stellate cells), RPTEC/TERT-1 (human proximal tubule) | [68] |
| PDMS-based device | OoC | Hepatocytes from primary and iPS-derived cells | [69] |
| Three-layered glass device | OoC, disease model, drug screening | Primary human hepatocytes, LSECs (Liver Sinusoidal Endothelial Cells), Kupffer cells (liver) | [70] |
| Three-layered glass device | OoC, disease model, drug screening | Primary human hepatocytes, iPSC (induced-Pluripotent Stem Cells), endothelial cells, Kupffer cells (liver) | [71] |
| Silicon scaffold fabricated by deep reactive ion etching | OoC, disease model, drug screening | PHH (Primary Human Hepatocyte), non-parenchymal cells | [72] |
| PDMS “open-top” device | Angiogenesis, spheroid formation | HDMEC (Human Dermal Micro-vascular Endothelial Cells), Primary human lung fibroblasts, U87MG (nervous tissue) | [73] |
| PDMS based device | Angiogenesis, OoC | hLFs (human Lung Fibroblasts), HUVECs (Human Umbilical Vein Endothelial Cells) | [74] |
| Two-layered glass-PDMS hybrid system | Spheroid formation, invasion assay, drug screening | U87 (nervous tissue) | [75] |
| 3D-printed system (Vero White Plus FullCure 835 resin) |
Angiogenesis, cell culture, drug screening | bEnd.3 (mouse brain endothelial cell line) | [76] |
| Double-casting of PDMS, with master mold made of PMMA. | Spheroid formation, drug screening | Caco-2 (Colon), NHDF (Normal Human Dermal Fibroblast), HepG2 (liver), A549 (lung) | [77] |
| 3D-hydrogel device | Drug screening, OoC | hCMEC/D3 (endothelial cell), HUVECs (Human Umbilical Vein Endothelial Cells), primary neurons, astrocytes | [78] |
| PDMS based device | OoC, drug screening | C3A (liver), EA.hy926 (endothelial) | [79] |
| PMMA-PDMS hybrid system and bioprinted hydrogel scaffold | OoC, angiogenesis | HUVECs (Human Umbilical Vein Endothelial Cells), neonatal rate cardiomyocytes | [80] |
| PDMS based device | OoC, disease model, drug screening | hiPSCs (human induced Pluripotent Stem Cells), CMs (Cardiomyocytes) differentiated from hiPSCs | [81] |
References
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33.
- Eder, C.; Falkner, E.; Nehrer, S.; Losert, U.M.; Schoeffl, H. Introducing the concept of the 3Rs into tissue engineering research. Altex 2006, 23, 17–23.
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures-a comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919.
- Ravi, M.; Paramesh, V.; Kaviya, S.R.; Anuradha, E.; Solomon, F.D. 3D cell culture systems: Advantages and applications. J. Cell. Physiol. 2015, 230, 16–26.
- Li, X.J.; Valadez, A.V.; Zuo, P.; Nie, Z. Microfluidic 3D cell culture: Potential application for tissue-based bioassays. Bioanalysis 2012, 4, 1509–1525.
- Toh, Y.C.; Lim, T.C.; Tai, D.; Xiao, G.; van Noort, D.; Yu, H. A microfluidic 3D hepatocyte chip for drug toxicity testing. Lab Chip 2009, 9, 2026–2035.
- Carrion, B.; Huang, C.P.; Ghajar, C.M.; Kachgal, S.; Kniazeva, E.; Jeon, N.L.; Putnam, A.J. Recreating the perivascular niche ex vivo using a microfluidic approach. Biotechnol. Bioeng. 2010, 107, 1020–1028.
- Cuchiara, M.P.; Allen, A.C.B.; Chen, T.M.; Miller, J.S.; West, J.L. Multilayer microfluidic PEGDA hydrogels. Biomaterials 2010, 31, 5491–5497.
- Choi, J.; Kim, S.; Jung, J.; Lim, Y.; Kang, K.; Park, S.; Kang, S. Wnt5a-mediating neurogenesis of human adipose tissue-derived stem cells in a 3D microfluidic cell culture system. Biomaterials 2011, 32, 7013–7022.
- Locatelli, L.; Inglebert, M.; Scrimieri, R.; Sinha, P.K.; Zuccotti, G.V.; Milani, P.; Bureau, L.; Misbah, C.; Maier, J.A.M. Human endothelial cells in high glucose: New clues from culture in 3D microfluidic chips. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2022, 36, e22137.
- Jachter, S.L.; Simmons, W.P.; Estill, C.; Xu, J.; Bishop, C.V. Matrix-free three-dimensional culture of bovine secondary follicles to antral stage: Impact of media formulation and epidermal growth factor (EGF). Theriogenology 2022, 181, 89–94.
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9.
- Wang, H.; Brown, P.C.; Chow, E.C.Y.; Ewart, L.; Ferguson, S.S.; Fitzpatrick, S.; Freedman, B.S.; Guo, G.L.; Hedrich, W.; Heyward, S.; et al. 3D cell culture models: Drug pharmacokinetics, safety assessment, and regulatory consideration. Clin. Transl. Sci. 2021, 14, 1659–1680.
- Olgasi, C.; Cucci, A.; Follenzi, A. iPSC-Derived Liver Organoids: A Journey from Drug Screening, to Disease Modeling, Arriving to Regenerative Medicine. Int. J. Mol. Sci. 2020, 21, 6215.
- Belfiore, L.; Aghaei, B.; Law, A.M.K.; Dobrowolski, J.C.; Raftery, L.J.; Tjandra, A.D.; Yee, C.; Piloni, A.; Volkerling, A.; Ferris, C.J.; et al. Generation and analysis of 3D cell culture models for drug discovery. Eur. J. Pharm. Sci. 2021, 163, 105876.
- Bahrami, S.; Baheiraei, N.; Najafi-Ashtiani, M.; Nour, S.; Razavi, M. Chapter 9-Microfluidic devices in tissue engineering. In Biomedical Applications of Microfluidic Devices; Hamblin, M.R., Karimi, M., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 209–233.
- Bahmaee, H.; Owen, R.; Boyle, L.; Perrault, C.M.; Garcia-Granada, A.A.; Reilly, G.C.; Claeyssens, F. Design and Evaluation of an Osteogenesis-on-a-Chip Microfluidic Device Incorporating 3D Cell Culture. Front. Bioeng. Biotechnol. 2020, 8.
- Choi, N.W.; Cabodi, M.; Held, B.; Gleghorn, J.P.; Bonassar, L.J.; Stroock, A.D. Microfluidic scaffolds for tissue engineering. Nat. Mater. 2007, 6, 908–915.
- Wu, Q.; Liu, J.; Wang, X.; Feng, L.; Wu, J.; Zhu, X.; Wen, W.; Gong, X. Organ-on-a-chip: Recent breakthroughs and future prospects. BioMed. Eng. OnLine 2020, 19, 9.
- van Duinen, V.; Trietsch, S.J.; Joore, J.; Vulto, P.; Hankemeier, T. Microfluidic 3D cell culture: From tools to tissue models. Curr. Opin. Biotechnol. 2015, 35, 118–126.
- van Engeland, N.C.A.; Pollet, A.; den Toonder, J.M.J.; Bouten, C.V.C.; Stassen, O.; Sahlgren, C.M. A biomimetic microfluidic model to study signalling between endothelial and vascular smooth muscle cells under hemodynamic conditions. Lab Chip 2018, 18, 1607–1620.
- Farhat, J.; Pandey, I.; AlWahsh, M. Transcending toward Advanced 3D-Cell Culture Modalities: A Review about an Emerging Paradigm in Translational Oncology. Cells 2021, 10, 1657.
- Nii, T.; Makino, K.; Tabata, Y. Three-Dimensional Culture System of Cancer Cells Combined with Biomaterials for Drug Screening. Cancers 2020, 12, 2754.
- Nii, T.; Makino, K.; Tabata, Y. A Cancer Invasion Model Combined with Cancer-Associated Fibroblasts Aggregates Incorporating Gelatin Hydrogel Microspheres Containing a p53 Inhibitor. Tissue Eng. Part C Methods 2019, 25, 711–720.
- De Waele, J.; Reekmans, K.; Daans, J.; Goossens, H.; Berneman, Z.; Ponsaerts, P. 3D culture of murine neural stem cells on decellularized mouse brain sections. Biomaterials 2015, 41, 122–131.
- Porzionato, A.; Stocco, E.; Barbon, S.; Grandi, F.; Macchi, V.; De Caro, R. Tissue-Engineered Grafts from Human Decellularized Extracellular Matrices: A Systematic Review and Future Perspectives. Int. J. Mol. Sci. 2018, 19, 4117.
- Grün, C.; Altmann, B.; Gottwald, E. Advanced 3D Cell Culture Techniques in Micro-Bioreactors, Part I: A Systematic Analysis of the Literature Published between 2000 and 2020. Processes 2020, 8, 1656.
- Manfredonia, C.; Muraro, M.G.; Hirt, C.; Mele, V.; Governa, V.; Papadimitropoulos, A.; Däster, S.; Soysal, S.D.; Droeser, R.A.; Mechera, R.; et al. Maintenance of Primary Human Colorectal Cancer Microenvironment Using a Perfusion Bioreactor-Based 3D Culture System. Adv. Biosyst. 2019, 3, e1800300.
- Priyadarshini, B.M.; Dikshit, V.; Zhang, Y. 3D-printed Bioreactors for In Vitro Modeling and Analysis. Int. J. Bioprint. 2020, 6, 267.
- Khan, I.; Prabhakar, A.; Delepine, C.; Tsang, H.; Pham, V.; Sur, M. A low-cost 3D printed microfluidic bioreactor and imaging chamber for live-organoid imaging. Biomicrofluidics 2021, 15, 024105.
- Haycock, J.W. 3D cell culture: A review of current approaches and techniques. Methods Mol. Biol. 2011, 695, 1–15.
- Myers, D.R.; Lam, W.A. Vascularized Microfluidics and Their Untapped Potential for Discovery in Diseases of the Microvasculature. Annu Rev. Biomed. Eng. 2021, 23, 407–432.
- Freitas, R.A. Nanomedicine, Volume I: Basic Capabilities; Landes Bioscience: Georgetown, TX, USA, 1999; Volume 1.
- Batra, S.; Rakusan, K. Capillary length, tortuosity, and spacing in rat myocardium during cardiac cycle. Am. J. Physiol. 1992, 263, H1369–H1376.
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Blood Vessels and Endothelial Cells. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002.
- Sarin, H. Physiologic upper limits of pore size of different blood capillary types and another perspective on the dual pore theory of microvascular permeability. J. Angiogenesis Res. 2010, 2, 14.
- Bulger, R.E.; Eknoyan, G.; Purcell, D.J., 2nd; Dobyan, D.C. Endothelial characteristics of glomerular capillaries in normal, mercuric chloride-induced, and gentamicin-induced acute renal failure in the rat. J. Clin. Investig. 1983, 72, 128–141.
- Tsai, H.F.; Trubelja, A.; Shen, A.Q.; Bao, G. Tumour-on-a-chip: Microfluidic models of tumour morphology, growth and microenvironment. J. R Soc. Interface 2017, 14, 20170137.
- Enderle, J.D. Chapter 7-Compartmental Modeling. In Introduction to Biomedical Engineering, 3rd ed.; Enderle, J.D., Bronzino, J.D., Eds.; Academic Press: Boston, MA, USA, 2012; pp. 359–445.
- Lipatov, V.A.; Kryukov, A.A.; Severinov, D.A.; Saakyan, A.R. Ethical and legal aspects of in vivo experimental biomedical research of the conduct. Part II. IP Pavlov Russ. Med. Biol. Her. 2019, 27, 245–257.
- Baumans, V. Use of animals in experimental research: An ethical dilemma? Gene Ther. 2004, 11, S64–S66.
- Mollaki, V. Ethical Challenges in Organoid Use. BioTech 2021, 10, 12.
- Bédard, P.; Gauvin, S.; Ferland, K.; Caneparo, C.; Pellerin, È.; Chabaud, S.; Bolduc, S. Innovative Human Three-Dimensional Tissue-Engineered Models as an Alternative to Animal Testing. Bioengineering 2020, 7, 115.
- Limongi, T.; Tirinato, L.; Pagliari, F.; Giugni, A.; Allione, M.; Perozziello, G.; Candeloro, P.; Di Fabrizio, E. Fabrication and Applications of Micro/Nanostructured Devices for Tissue Engineering. Nano-Micro Lett. 2016, 9, 1.
- Limongi, T.; Cesca, F.; Gentile, F.; Marotta, R.; Ruffilli, R.; Barberis, A.; Dal Maschio, M.; Petrini, E.M.; Santoriello, S.; Benfenati, F.; et al. Nanostructured superhydrophobic substrates trigger the development of 3D neuronal networks. Small 2013, 9, 402–412.
- Limongi, T.; Rocchi, A.; Cesca, F.; Tan, H.; Miele, E.; Giugni, A.; Orlando, M.; Perrone Donnorso, M.; Perozziello, G.; Benfenati, F.; et al. Delivery of Brain-Derived Neurotrophic Factor by 3D Biocompatible Polymeric Scaffolds for Neural Tissue Engineering and Neuronal Regeneration. Mol. Neurobiol. 2018, 55, 8788–8798.
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells 2019, 11, 1065–1083.
- Gu, C.; Feng, J.; Waqas, A.; Deng, Y.; Zhang, Y.; Chen, W.; Long, J.; Huang, S.; Chen, L. Technological Advances of 3D Scaffold-Based Stem Cell/Exosome Therapy in Tissues and Organs. Front. Cell Dev. Biol. 2021, 9, 709204.
- Zhang, L.; Yang, G.; Johnson, B.N.; Jia, X. Three-dimensional (3D) printed scaffold and material selection for bone repair. Acta Biomater. 2019, 84, 16–33.
- Dattola, E.; Parrotta, E.I.; Scalise, S.; Perozziello, G.; Limongi, T.; Candeloro, P.; Coluccio, M.L.; Maletta, C.; Bruno, L.; De Angelis, M.T.; et al. Development of 3D PVA scaffolds for cardiac tissue engineering and cell screening applications. RSC Adv. 2019, 9, 4246–4257.
- Tong, A.; Voronov, R. A Minireview of Microfluidic Scaffold Materials in Tissue Engineering. Front. Mol. Biosci. 2022, 8, 783268.
- Currens, E.R.; Armbruster, M.R.; Castiaux, A.D.; Edwards, J.L.; Martin, R.S. Evaluation and optimization of PolyJet 3D-printed materials for cell culture studies. Anal. Bioanal Chem 2022, 414, 3329–3339.
- Ma, L.-D.; Wang, Y.-T.; Wang, J.-R.; Wu, J.-L.; Meng, X.-S.; Hu, P.; Mu, X.; Liang, Q.-L.; Luo, G.-A. Design and fabrication of a liver-on-a-chip platform for convenient, highly efficient, and safe in situ perfusion culture of 3D hepatic spheroids. Lab Chip 2018, 18, 2547–2562.
- Lin, Z.; Rao, Z.; Chen, J.; Chu, H.; Zhou, J.; Yang, L.; Quan, D.; Bai, Y. Bioactive Decellularized Extracellular Matrix Hydrogel Microspheres Fabricated Using a Temperature-Controlling Microfluidic System. ACS Biomater. Sci. Eng. 2022, 8, 1644–1655.
- Yu, J.; Lee, S.; Song, J.; Lee, S.R.; Kim, S.; Choi, H.; Kang, H.; Hwang, Y.; Hong, Y.K.; Jeon, N.L. Perfusable micro-vascularized 3D tissue array for high-throughput vascular phenotypic screening. Nano Converg. 2022, 9, 16.
- Shin, W.; Kim, H.J. 3D in vitro morphogenesis of human intestinal epithelium in a gut-on-a-chip or a hybrid chip with a cell culture insert. Nat. Protoc. 2022, 17, 910–939.
- Kumon, H.; Sakuma, S.; Nakamura, S.; Maruyama, H.; Eto, K.; Arai, F. Microfluidic Bioreactor Made of Cyclo-Olefin Polymer for Observing On-Chip Platelet Production. Micromachines 2021, 12, 1253.
- Carnero, B.; Bao-Varela, C.; Gómez-Varela, A.I.; Álvarez, E.; Flores-Arias, M.T. Microfluidic devices manufacturing with a stereolithographic printer for biological applications. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 129, 112388.
- Ahn, J.; Lim, J.; Jusoh, N.; Lee, J.; Park, T.-E.; Kim, Y.; Kim, J.; Jeon, N.L. 3D Microfluidic Bone Tumor Microenvironment Comprised of Hydroxyapatite/Fibrin Composite. Front. Bioeng. Biotechnol. 2019, 7, 168.
- Vernetti, L.A.; Senutovitch, N.; Boltz, R.; DeBiasio, R.; Shun, T.Y.; Gough, A.; Taylor, D.L. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp. Biol. Med. 2016, 241, 101–114.
- Shim, K.Y.; Lee, D.; Han, J.; Nguyen, N.T.; Park, S.; Sung, J.H. Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed. Microdevices 2017, 19, 37.
- Mosadegh, B.; Lockett, M.R.; Minn, K.T.; Simon, K.A.; Gilbert, K.; Hillier, S.; Newsome, D.; Li, H.; Hall, A.B.; Boucher, D.M.; et al. A paper-based invasion assay: Assessing chemotaxis of cancer cells in gradients of oxygen. Biomaterials 2015, 52, 262–271.
- Bircsak, K.M.; DeBiasio, R.; Miedel, M.; Alsebahi, A.; Reddinger, R.; Saleh, A.; Shun, T.; Vernetti, L.A.; Gough, A. A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate®. Toxicology 2021, 450, 152667.
- Park, D.; Son, K.; Hwang, Y.; Ko, J.; Lee, Y.; Doh, J.; Jeon, N.L. High-Throughput Microfluidic 3D Cytotoxicity Assay for Cancer Immunotherapy (CACI-IMPACT Platform). Front. Immunol. 2019, 10, 1133.
- Ong, L.J.Y.; Islam, A.; DasGupta, R.; Iyer, N.G.; Leo, H.L.; Toh, Y.C. A 3D printed microfluidic perfusion device for multicellular spheroid cultures. Biofabrication 2017, 9, 045005.
- Chen, J.; Liu, C.Y.; Wang, X.; Sweet, E.; Liu, N.; Gong, X.; Lin, L. 3D printed microfluidic devices for circulating tumor cells (CTCs) isolation. Biosens. Bioelectron. 2020, 150, 111900.
- Lee, J.; Choi, B.; No da, Y.; Lee, G.; Lee, S.R.; Oh, H.; Lee, S.H. A 3D alcoholic liver disease model on a chip. Integr. Biol. Quant. Biosci. Nano Macro 2016, 8, 302–308.
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Hübner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A.; et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015, 15, 2688–2699.
- Schepers, A.; Li, C.; Chhabra, A.; Seney, B.T.; Bhatia, S. Engineering a perfusable 3D human liver platform from iPS cells. Lab Chip 2016, 16, 2644–2653.
- Li, X.; George, S.M.; Vernetti, L.; Gough, A.H.; Taylor, D.L. A glass-based, continuously zonated and vascularized human liver acinus microphysiological system (vLAMPS) designed for experimental modeling of diseases and ADME/TOX. Lab Chip 2018, 18, 2614–2631.
- Sakolish, C.; Reese, C.E.; Luo, Y.S.; Valdiviezo, A.; Schurdak, M.E.; Gough, A.; Taylor, D.L.; Chiu, W.A.; Vernetti, L.A.; Rusyn, I. Analysis of reproducibility and robustness of a human microfluidic four-cell liver acinus microphysiology system (LAMPS). Toxicology 2021, 448, 152651.
- Ortega-Prieto, A.M.; Skelton, J.K.; Wai, S.N.; Large, E.; Lussignol, M.; Vizcay-Barrena, G.; Hughes, D.; Fleck, R.A.; Thursz, M.; Catanese, M.T.; et al. 3D microfluidic liver cultures as a physiological preclinical tool for hepatitis B virus infection. Nat. Commun. 2018, 9, 682.
- Oh, S.; Ryu, H.; Tahk, D.; Ko, J.; Chung, Y.; Lee, H.K.; Lee, T.R.; Jeon, N.L. “Open-top” microfluidic device for in vitro three-dimensional capillary beds. Lab Chip 2017, 17, 3405–3414.
- Nashimoto, Y.; Hayashi, T.; Kunita, I.; Nakamasu, A.; Torisawa, Y.-s.; Nakayama, M.; Takigawa-Imamura, H.; Kotera, H.; Nishiyama, K.; Miura, T.; et al. Integrating perfusable vascular networks with a three-dimensional tissue in a microfluidic device. Integr. Biol. 2017, 9, 506–518.
- Ma, J.; Li, N.; Wang, Y.; Wang, L.; Wei, W.; Shen, L.; Sun, Y.; Jiao, Y.; Chen, W.; Liu, J. Engineered 3D tumour model for study of glioblastoma aggressiveness and drug evaluation on a detachably assembled microfluidic device. Biomed. Microdevices 2018, 20, 80.
- Kim, J.A.; Kim, H.N.; Im, S.-K.; Chung, S.; Kang, J.Y.; Choi, N. Collagen-based brain microvasculature model in vitro using three-dimensional printed template. Biomicrofluidics 2015, 9, 024115.
- Eilenberger, C.; Rothbauer, M.; Selinger, F.; Gerhartl, A.; Jordan, C.; Harasek, M.; Schädl, B.; Grillari, J.; Weghuber, J.; Neuhaus, W.; et al. A Microfluidic Multisize Spheroid Array for Multiparametric Screening of Anticancer Drugs and Blood–Brain Barrier Transport Properties. Adv. Sci. 2021, 8, 2004856.
- Adriani, G.; Ma, D.; Pavesi, A.; Kamm, R.D.; Goh, E.L.K. A 3D neurovascular microfluidic model consisting of neurons, astrocytes and cerebral endothelial cells as a blood–brain barrier. Lab Chip 2017, 17, 448–459.
- Jia, Z.; Cheng, Y.; Jiang, X.; Zhang, C.; Wang, G.; Xu, J.; Li, Y.; Peng, Q.; Gao, Y. 3D Culture System for Liver Tissue Mimicking Hepatic Plates for Improvement of Human Hepatocyte (C3A) Function and Polarity. BioMed Res. Int. 2020, 2020, 6354183.
- Zhang, Y.S.; Arneri, A.; Bersini, S.; Shin, S.R.; Zhu, K.; Goli-Malekabadi, Z.; Aleman, J.; Colosi, C.; Busignani, F.; Dell’Erba, V.; et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016, 110, 45–59.
- Veldhuizen, J.; Nikkhah, M. Developing 3D Organized Human Cardiac Tissue within a Microfluidic Platform. J. Vis. Exp. JoVE 2021.





