Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Nadia Halib | -- | 1962 | 2022-05-20 05:54:16 | | | |
| 2 | Jessie Wu | -2 word(s) | 1960 | 2022-05-20 06:05:09 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Halib, N.; Grassi, G.; Pavan, N.; Trombetta, C.; Dapas, B.; , .; Scaggiante,, B.; Grassi, M. siRNA Structure, Function and Delivery. Encyclopedia. Available online: https://encyclopedia.pub/entry/23151 (accessed on 07 February 2026).
Halib N, Grassi G, Pavan N, Trombetta C, Dapas B, , et al. siRNA Structure, Function and Delivery. Encyclopedia. Available at: https://encyclopedia.pub/entry/23151. Accessed February 07, 2026.
Halib, Nadia, Gabriele Grassi, Nicola Pavan, Carlo Trombetta, Barbara Dapas, , Bruna Scaggiante,, Mario Grassi. "siRNA Structure, Function and Delivery" Encyclopedia, https://encyclopedia.pub/entry/23151 (accessed February 07, 2026).
Halib, N., Grassi, G., Pavan, N., Trombetta, C., Dapas, B., , ., Scaggiante,, B., & Grassi, M. (2022, May 20). siRNA Structure, Function and Delivery. In Encyclopedia. https://encyclopedia.pub/entry/23151
Halib, Nadia, et al. "siRNA Structure, Function and Delivery." Encyclopedia. Web. 20 May, 2022.
Copy Citation
siRNA is a double-stranded RNA molecule with 21- and 22-nucleotide generated by ribonuclease III cleavage from longer double-stranded RNA (dsRNAs). After binding to the RNA-induced silencing complex (RISC) in the cytoplasm, the sense strand of siRNA undergoes ejection, while the antisense strand of siRNA targets the complementary messenger RNA (mRNA).
siRNA
cancer
liposomes
1. Introduction
Partial hybridization of the antisense strand of siRNA with the target mRNA leads to inhibition of translation, while perfect complementary hybridization causes degradation of the mRNA (Figure 1). Thus, siRNA can effectively down-regulate gene expression. Notably, siRNA can be chemically synthesized to target virtually any mRNA of disease-causing gene; thus, they have the potential to be used as a therapeutic agent in many human diseases including urological cancers.
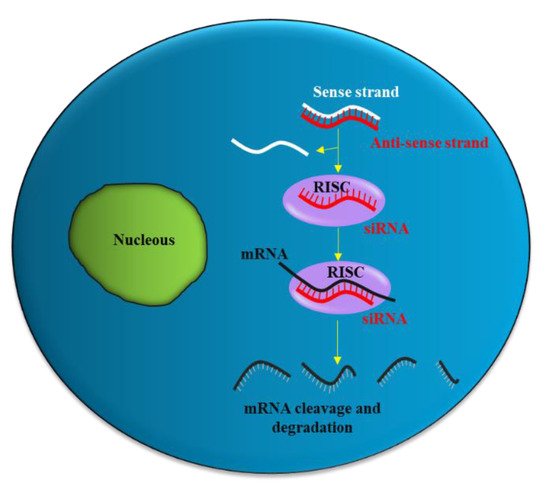
Figure 1. siRNA mechanism of action. The antisense strand (red) of the siRNA is taken up by a catalytic protein complex (RNA-induced silencing complex, RISC), and the sense strand (white) of the siRNA is discarded. The antisense strand drives RISC to a target mRNA (black), which results in specific, RISC-mediated mRNA degradation.
2. siRNA Delivery Problems
Like all nucleic acid-based molecules [1][2][3], siRNA cannot enter cells on their own and require a delivery agent [4][5][6][7]. When delivered systemically, naked siRNAs have to deal with different biological barriers. In particular, they can: (1) Be degraded by blood nucleases; (2) be eliminated by the phagocytic system, (3) be sequestered by the liver and filtrated by kidney [8] (Figure 2). Additionally, siRNAs have to face the problem of blood wall crossing (extravasation) (4) and migration through the extracellular matrix (5). When they reach the target cell, they cannot efficiently cross the cell membrane (6). Indeed, the presence of phosphate groups in their structure confers to siRNAs a global negative electrical charge that hinders the interaction with the negatively charged surface of the cells, which tends to repulse them. Moreover, siRNA hydrophilic nature prevents the crossing through the hydrophobic inner layer of the cell membrane. Once in the target cell, endosomal escape [9] has to be accomplished (7). If sequestered into endosomes, siRNAs do not have the possibility to get in contact with their targets, thus drastically impairing if not abolishing the biological effect(s). All these obstacles may eventually lead to a negligible effectiveness for siRNA.
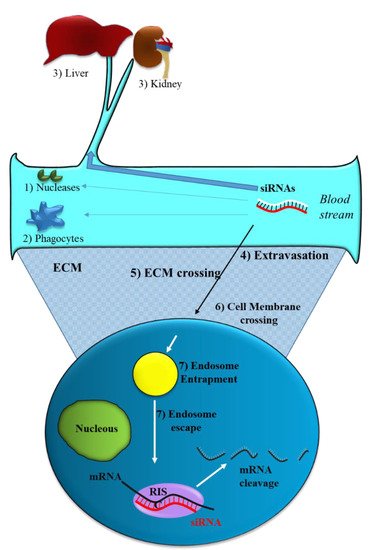
Figure 2. Obstacles to siRNA delivery. Systemically released siRNAs encounter blood nucleases (1), which can induce their rapid degradation together with the clearance by phagocytes (2) and by the liver–kidney sequestration/filtration (3). Extravasation (4), extra cellular matrix (ECM) crossing (5), cell membrane crossing (6) and endosomal escape (7) are the other barriers to be overcome by siRNAs.
Beside the above aspects, another feature of tumor tissue can be considered to optimize siRNA delivery. The aberrant tumor neo-vasculature is responsible for ineffective oxygen delivery in the inner tumor regions. Thus, tumor cells can derive their energy mostly from anaerobic glycolysis, which determines the increased production of lactic acid. This, in concomitance with the reduced H + removal by the defective neo-vasculature, causes the reduction of tumor tissue pH. This feature may be considered to generate delivery systems that preferentially release siRNA in low pH [10]. By appropriately taking into account all the above aspects, it is in principle possible to minimize the negative effects of the bio-barriers on siRNA delivery improving the effectiveness of these molecules in the biological environment.
3. Strategies to Optimize siRNA Delivery
A well-recognized strategy to promote siRNA delivery is based on the use of synthetic vectors that can protect siRNA and potentially deliver it to target cells. Several synthetic materials have been used for delivery of siRNAs as briefly reported below and summarized in Table 1.
Table 1. Properties of siRNA delivery materials.
| Delivery Material | Advantages | Disadvantages |
|---|---|---|
| Liposomes | Easy siRNA loading Minor toxicity Structure easily tunable |
No targeting specificity unless equipped with targeting moieties |
| Echogenic liposomes | As above with the possibility to induce ultrasound controlled delivery | Not applicable in deep tissue |
| Exosomes | As above with excellent biodistribution and the possibility to escape clearance by the mononuclear phagocyte system | No targeting specificity |
| Polymers | Production/isolation non expensive Structure easily tunable In general non toxic Possibility to escape endosome (PEI) In general easy siRNA loading Targeting ability to CD44 (HA) |
Described toxicity for PEI Low solubility for CH Electrostatic repulsion of siRNA due to polyanionic nature for HA |
| Aptamers | Non toxic Able to target any desired molecule For DNA aptamers low production cost Can easily be stored for very long periods without losing their activity |
RNA aptamers may be unstable in the biological environment. The selection procedure may be complex |
| Magnetic nanoparticles | Large surface area which can be functionalized with smart functional groups Magnetic behavior allows targeting to a defined tisse following application of an external magnetic field |
Need functional groups on their surface for siRNA loading |
| Carbon nanotubes | High drug loading capacity Cellular uptake can be modulated varying the dimension |
Poorly biodegradable |
PEI: polyethylenimine; CH: chitosan; CD44: cluster determinant 44; HA: Hyaluronic acid.
4. Lipid-Based Delivery Materials
The most well known type of lipid-based delivery material is liposomes (Figure 3A). These lipid particles contain an inner aqueous space separated from the outer environment by a bilayer membrane composed of amphiphilic lipids. The hydrophilic heads of the amphiphilic lipids are oriented towards the outer and inner environment; the hydrophobic tails form a hydrophobic environment inside the membrane. A cationic head group characterizes the amphiphilic lipids used for siRNA delivery; this group allows the interaction with the negatively charged phosphate groups present in the siRNA structure.
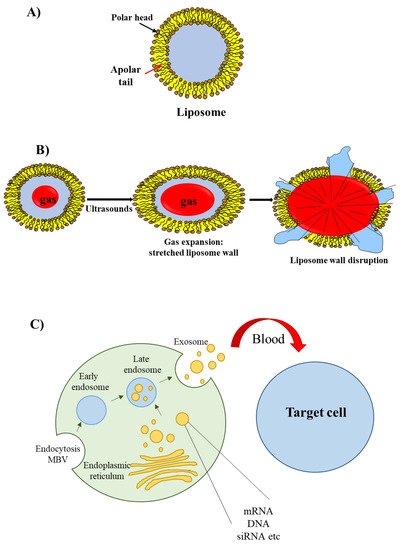
Figure 3. Liposomes, echogenic liposomes and exosomes. (A) The liposome wall consists of a double layer of amphiphilic lipids containing a polar head (oriented either towards the external or internal environments) and a polar tail (oriented towards the lipophilic region of the membrane). (B) Echogenic liposomes contain an inner gas phase that, in the presence of ultrasounds, undergoes expansion, contraction and vibration (cavitation); this phenomenon eventually determines liposome wall stretching and rupture. (C) Exosomes originate from early and late endosomes, which derive from invagination of the limited multi-vesicular body membrane (MBV); during this process, cellular proteins, mRNA, miRNA and DNA fragment are incorporated within the exosome; following exosome delivery form the mother cell, cellular proteins/mRNA/miRNA/DNA fragment are released to recipient cells.
An interesting group of liposomes are the echogenic liposomes [11]. These are ordinary liposomes, which contain a gas in their internal aqueous environment (Figure 3B). When exposed to ultrasounds consisting of pressure waves with frequencies equal or greater than 20 kHz, echogenic liposomes undergo the phenomenon of cavitation. Cavitation consists of the rapid growth and collapse of bubbles or the sustained oscillatory movement of bubbles. This phenomenon favors the release of siRNA [12] as cavitation causes the formation of transient pores in the cellular membrane that favors the uptake of siRNAs [13].
Recently, a novel class of lipid particles named exosomes has attracted the attention of researchers. These are extracellular nanovesicles with a size of 40–100 nm that are produced by various cells and released into the extracellular environment upon fusion with the plasma membrane. Exosomes arise from early and late endosomes (Figure 3C) that result from invagination of the limited multivesicular body (MVB) membrane [14]. As exosomes can be obtained from patients’ body fluids, they have excellent biodistribution and biocompatibility when reinjected into the same patient for drug delivery purposes [15].
As reported in Section 2.1, the pH responsive liposomes are of interest for the tumor-targeted delivery of siRNAs. Usually they contain a neutral lipid and a weakly acidic amphiphile [16]. In the acidic environment, the negatively charged group of the phospholipid is destabilized, eventually favoring fusion with the cell membrane or with the endosomal membrane and siRNA release. A recent approach to generating pH sensitive liposomes is based on the conjugation with pH-responsive polymers [17].
5. Polymer-Based Delivery Materials
Polymers are commonly used for siRNA delivery because they are not expensive to produce/isolate and can be easily modified. Typically, they contain cationic groups that allow electrostatic interaction with the negatively charged siRNAs. A widely used polymer for siRNA delivery is polyethylenimine (PEI) [18] that promotes the escape of siRNA from endosome, thereby enhancing the cytosolic availability of siRNAs [19]. Since it displays some toxicity, it is often conjugated with poly(ethylene glycol) (PEG), which has the ability to reduce cytotoxicity and improve stability in the presence of serum proteins [20].
PLGA is a copolymer of polylactic acid (PLA) and polyglycolic acid (PGA) with a variable number of lactic and glycol acid units [21]. It is an FDA-approved polymer that is physically strong and biocompatible/biodegradable.
Chitosan (CH) is obtained by deacetylation of chitin, which is found in the exoskeleton of crustaceans and in the cell walls of fungi. It is a linear polysaccharide with a carbohydrate backbone containing two types of repeating residues, 2-amino-2-deoxy-glucose (glucosamine) and 2-N-acetyl-2-deoxy-glucose (N-glucosamine) [22]. The amino groups give CH a positive charge that enables electrostatic interaction with siRNAs.
Hyaluronic acid (HA) is a linear polysaccharide [23] that, due to the presence of HA receptors in most cancer tissues, has a great potential for targeted drug delivery. In particular, it binds cluster determinant 44 (CD44), an adhesion molecule that is highly expressed in a variety of tumor cells.
Polymers have also been used to create special structures called dendrimers (from the Greek “dendron” (tree) and “meros” (part)). Dendrimers have a central core molecule from which tree-like arms extend in an orderly symmetrical manner [24]. The arms are organized in different layers and the complexity of the dendrimer depends on the number of layers. For siRNA delivery, dendrimers are usually made of polymers that confer a net cationic surface charge to the structure; this allows the siRNA to bind via electrostatic interactions.
The generation of pH responsive polymers can be of great interest for the tumor specific delivery of siRNAs. In this regard, there are two main types of pH responsive polymers [25]: Those with ionizable moieties and those containing acid-labile linkages. In the first case, ionizable moieties such as amines and carboxylic acids are protonated or deprotonated in relation to the pH. The protonation/deprotonation phenomenon determines structural changes in the polymer which allows the release of the drug. In the case of tumor tissue, the decreased pH induces polymer protonation, thus triggering siRNA delivery. Many different polymers can be used for this purpose as long as they contain ionisable moieties. In the second type of pH responsive polymers, the polymer backbone contains acid-labile covalent linkages that are cleaved due to pH decrease. This results in the polymer degradation with the consequent siRNA delivery at the site of increase acidity, i.e, the inner tumor tissue.
6. Other Delivery Materials
Nucleic acid Aptamers are short single stranded non-coding DNA or RNA molecules [26]. They form secondary/tertiary structures, which eventually determines their 3D shape, responsible for their ability to bind specifically to a large number of target biological molecules. In the biomedical field, aptamers are used as drugs per se [27] or to decorate nanoparticles (of any material) to target specific cellular antigens.
Iron oxide based magnetic nanoparticles (IONPs) are composed of magnetic iron oxides, elements that are widely distributed in nature and easily synthesized in the laboratory. IONPs have a large surface area and can be engineered with functional groups to allow cross-linking with monoclonal antibodies, peptides or small molecules (such as aptamers) for both diagnostic imaging and therapeutic purposes [28]. The major advantage of IONPs in drug delivery is that once the particles enter the blood, they can be recruited to a specific body site by applying an external high-gradient magnetic field.
Carbon nanotubes (CNTs) are an emerging and attractive delivery material for siRNAs [29]. CNTs can be in the form of single-walled carbon nanotubes with a cylindrical shape or in the form of multi-layer graphene sheets wrapped around each other in a cylindrical shape. CNTs are of potential interest for biomedicine and especially for drug/siRNA delivery due to their properties such as large surface area, flexible interaction with cargo, high drug loading capacity and ability to release therapeutic agents at target sites. However, the lack of biodegradability and toxicity has so far limited their full potential in the biomedical field.
References
- Grassi, M.; Cavallaro, G.; Scirè, S.; Scaggiante, B.; Daps, B.; Farra, R.; Baiz, D.; Giansante, C.; Guarnieri, G.; Perin, D.; et al. Current strategies to improve the efficacy and the delivery of nucleic acid based drugs. Curr. Signal Transduct. Ther. 2010, 5, 92–120.
- Grassi, G.; Dawson, P.; Guarnieri, G.; Kandolf, R.; Grassi, M. Therapeutic potential of hammerhead ribozymes in the treatment of hyper-proliferative diseases. Curr. Pharm. Biotechnol. 2004, 5, 369–386.
- Grassi, G.; Marini, J.C. Ribozymes: Structure, function, and potential therapy for dominant genetic disorders. Ann. Med. 1996, 28, 499–510.
- Farra, R.; Musiani, F.; Perrone, F.; Cemazar, M.; Kamensek, U.; Tonon, F.; Abrami, M.; Rucigaj, A.; Grassi, M.; Pozzato, G.; et al. Polymer-mediated delivery of siRNAs to hepatocellular carcinoma: Variables affecting specificity and effectiveness. Molecules 2018, 23, 777.
- Scarabel, L.; Perrone, F.; Garziera, M.; Farra, R.; Grassi, M.; Musiani, F.; Russo, S.C.; Salis, B.; De, S.L.; Toffoli, G.; et al. Strategies to optimize siRNA delivery to hepatocellular carcinoma cells. Expert Opin. Drug Deliv. 2017, 14, 797–810.
- Farra, R.; Maruna, M.; Perrone, F.; Grassi, M.; Benedetti, F.; Maddaloni, M.; El, B.M.; Parisi, S.; Rizzolio, F.; Forte, G.; et al. Strategies for delivery of siRNAs to ovarian cancer cells. Pharmaceutics 2019, 11, 547.
- Barba, A.A.; Lamberti, G.; Sardo, C.; Dapas, B.; Abrami, M.; Grassi, M.; Farra, R.; Tonon, F.; Forte, G.; Musiani, F.; et al. Novel lipid and polymeric materials as delivery systems for nucleic acid based drugs. Curr. Drug Metab. 2015, 16, 427–452.
- Huang, Y.; Hong, J.; Zheng, S.; Ding, Y.; Guo, S.; Zhang, H.; Zhang, X.; Du, Q.; Liang, Z. Elimination pathways of systemically delivered siRNA. Mol. Ther. 2011, 19, 381–385.
- Kaneda, Y. Gene therapy: A battle against biological barriers. Curr. Mol. Med. 2001, 1, 493–499.
- Wang, M.; Li, J.; Li, X.; Mu, H.; Zhang, X.; Shi, Y.; Chu, Y.; Wang, A.; Wu, Z.; Sun, K. Magnetically and pH dual responsive dendrosomes for tumor accumulation enhanced folate-targeted hybrid drug delivery. J. Control. Release 2016, 232, 161–174.
- Huang, S.L. Liposomes in ultrasonic drug and gene delivery. Adv. Drug Deliv. Rev. 2008, 60, 1167–1176.
- Husseini, G.A.; Pitt, W.G. Micelles and nanoparticles for ultrasonic drug and gene delivery. Adv. Drug Deliv. Rev. 2008, 60, 1137–1152.
- Nyborg, W.L. Ultrasound, contrast agents and biological cells; a simplified model for their interaction during in vitro experiments. Ultrasound Med. Biol. 2006, 32, 1557–1568.
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977.
- Natasha, G.; Gundogan, B.; Tan, A.; Farhatnia, Y.; Wu, W.; Rajadas, J.; Seifalian, A.M. Exosomes as immunotheranostic nanoparticles. Clin. Ther. 2014, 36, 820–829.
- Nayar, R.; Schroit, A.J. Generation of pH-sensitive liposomes: Use of large unilamellar vesicles containing N-succinyldioleoylphosphatidylethanolamine. Biochemistry 1985, 24, 5967–5971.
- Yoshizaki, Y.; Yuba, E.; Komatsu, T.; Udaka, K.; Harada, A.; Kono, K. Improvement of peptide-based tumor immunotherapy using pH-sensitive fusogenic polymer-modified liposomes. Molecules 2016, 21, 1284.
- Park, T.G.; Jeong, J.H.; Kim, S.W. Current status of polymeric gene delivery systems. Adv. Drug Deliv. Rev. 2006, 58, 467–486.
- York, A.W.; Kirkland, S.E.; McCormick, C.L. Advances in the synthesis of amphiphilic block copolymers via RAFT polymerization: Stimuli-responsive drug and gene delivery. Adv. Drug Deliv. Rev. 2008, 60, 1018–1036.
- Roberts, M.J.; Bentley, M.D.; Harris, J.M. Chemistry for peptide and protein PEGylation. Adv. Drug Deliv. Rev. 2002, 54, 459–476.
- Makadia, H.K.; Siegel, S.J. Poly Lactic-co-glycolic acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers 2011, 3, 1377–1397.
- Serrano-Sevilla, I.; Artiga, A.; Mitchell, S.G.; De, M.L.; de la Fuente, J.M. Natural polysaccharides for siRNA delivery: Nanocarriers based on chitosan, hyaluronic acid, and their derivatives. Molecules 2019, 24, 2570.
- Dufes, C.; Uchegbu, I.F.; Schatzlein, A.G. Dendrimers in gene delivery. Adv. Drug Deliv. Rev. 2005, 57, 2177–2202.
- Tang, H.; Zhao, W.; Yu, J.; Li, Y.; Zhao, C. Recent development of pH-responsive polymers for cancer nanomedicine. Molecules 2018, 24, 4.
- Scaggiante, B.; Dapas, B.; Grassi, M.; Zanconati, F.; Farra, R.; Tonon, F.; Fiorentino, S.M.; Abrami, M.; Grassi, G. Nucleic acid-based aptamers and their applications. Future Med. 2013, 55–71.
- Dapas, B.; Pozzato, G.; Zorzet, S.; Capolla, S.; Paolo, M.; Scaggiante, B.; Coan, M.; Guerra, C.; Gnan, C.; Gattei, V.; et al. Effects of eEF1A1 targeting by aptamer/siRNA in chronic lymphocytic leukaemia cells. Int. J. Pharm. 2020, 574, 118895.
- Nedyalkova, M.; Donkova, B.; Romanova, J.; Tzvetkov, G.; Madurga, S.; Simeonov, V. Iron oxide nanoparticles—In vivo/in vitro biomedical applications and in silico studies. Adv. Colloid Interface Sci. 2017, 249, 192–212.
- Zare, H.; Ahmadi, S.; Ghasemi, A.; Ghanbari, M.; Rabiee, N.; Bagherzadeh, M.; Karimi, M.; Webster, T.J.; Hamblin, M.R.; Mostafavi, E. Carbon nanotubes: Smart drug/gene delivery carriers. Int. J. Nanomed. 2021, 16, 1681–1706.
- Nogawa, M.; Yuasa, T.; Kimura, S.; Tanaka, M.; Kuroda, J.; Sato, K.; Yokota, A.; Segawa, H.; Toda, Y.; Kageyama, S.; et al. Intravesical administration of small interfering RNA targeting PLK-1 successfully prevents the growth of bladder cancer. J. Clin. Investig. 2005, 115, 978–985.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
6.3K
Revisions:
2 times
(View History)
Update Date:
20 May 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




