You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Popov Roman | -- | 3269 | 2022-05-18 11:03:01 | | | |
| 2 | Amina Yu | -103 word(s) | 3166 | 2022-05-19 04:37:57 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Roman, P.; Ivanchina, N.V.; Dmitrenok, P. MS-Based Metabolomic Workflows in Marine Natural Products Analysis. Encyclopedia. Available online: https://encyclopedia.pub/entry/23060 (accessed on 22 December 2025).
Roman P, Ivanchina NV, Dmitrenok P. MS-Based Metabolomic Workflows in Marine Natural Products Analysis. Encyclopedia. Available at: https://encyclopedia.pub/entry/23060. Accessed December 22, 2025.
Roman, Popov, Natalia V. Ivanchina, Pavel Dmitrenok. "MS-Based Metabolomic Workflows in Marine Natural Products Analysis" Encyclopedia, https://encyclopedia.pub/entry/23060 (accessed December 22, 2025).
Roman, P., Ivanchina, N.V., & Dmitrenok, P. (2022, May 18). MS-Based Metabolomic Workflows in Marine Natural Products Analysis. In Encyclopedia. https://encyclopedia.pub/entry/23060
Roman, Popov, et al. "MS-Based Metabolomic Workflows in Marine Natural Products Analysis." Encyclopedia. Web. 18 May, 2022.
Copy Citation
Marine natural products are considered one of the main sources of compounds for drug development. Starfish and sea cucumbers are potential sources of natural products of pharmaceutical interest. Among their metabolites, polar steroids, triterpene glycosides, and polar lipids have attracted a great deal of attention.
starfish
sea cucumber
mass spectrometry
metabolomics
1. Introduction
The emergence of novel diseases and resistant forms of known diseases in recent years and the emergence of multidrug-resistant pathogens has led to a renewed interest in the exploration of new sources of bioactive compounds. The sea environment possesses extraordinary ecological variety, and its inhabitants exhibit enormous biochemical diversity. At present, over 29,000 marine natural products have been discovered [1][2][3][4]. To date, several dozen marine natural products or their derivatives have been approved as therapeutic agents or are undergoing Phase III, II, or I drug development [5]. As the biodiversity of marine organisms is higher than that of terrestrial plants and animals, and as only a minor portion of metabolites present in marine species has been studied, it can be assumed that the number of new marine compounds will continue to increase, providing new therapeutic alternatives.
Marine invertebrates have long been considered an inexhaustible source of novel natural products. Although Porifera and Cnidaria are the two major sources of new marine natural products, Echinodermata is viewed as another abundant source of new bioactive compounds. Over the past five years, just over two hundred new compounds have been isolated from echinoderms [1][2][3][4][6]. The phylum Echinodermata includes about 7500 species found in all seas at every depth, from intertidal to abyssal, and in all ecosystems, from coral reefs to shallow shores. Echinoderms are divided into five different taxonomic classes, including Asteroidea (starfish) and Holothuroidea (sea cucumbers).
Starfish and sea cucumbers are extensively employed in traditional medicine, being a rich source of bioactive compounds. Several starfish species are used to treat rheumatism or as tonics in traditional Chinese medicine [7][8]. Sea cucumbers are one of the most valuable aquaculture species in China, Korea, and Japan, as well as other countries, where they are used as functional foods. Traditional medicine in China and other countries in Asia and the Middle East uses sea cucumbers widely to treat a broad range of diseases, including asthma, arthritis, hypertension, and kidney disease [9][10].
Unique pharmacological properties, including anticancer, antioxidant, antithrombotic, and immunostimulating activities, among others, are associated with bioactive starfish and sea cucumber compounds [11][12][13][14][15]. Moreover, the secondary metabolites of sea cucumbers affect the biological clock and circadian rhythm of lipid metabolism [16], reduce fat accumulation [17], and protect against high fat diet-induced metabolic disorders in mice [18]. Extracts of certain specimens may accelerate wound healing and tissue regeneration [19].
The distinctive chemical composition of starfish and sea cucumbers seems to be the main reason for these beneficial properties. Starfish and sea cucumbers are high in valuable nutrients such as vitamins, minerals, and metabolites such as peptides, sterols, phenolics, sphingolipids, glycosaminoglycans, sulfated polysaccharides, and lectins [10]. Among these compounds the most exciting are unique polar steroid compounds and triterpene glycosides, which are characteristic of starfish and sea cucumbers. These compounds have unusual chemical structures and demonstrate a variety of biological effects, such as cytotoxic, antifungal, antiviral, antibacterial, anti-inflammatory, analgesic, ichthyotoxic, hemolytic, anti-biofouling, anticancer, immunomodulating, and neuritogenic actions [12][20][21][22][23][24][25][26][27][28][29][30][31][32][33].
Secondary metabolites are usually present in the extracts of starfish and sea cucumbers as complex mixtures of very similar compounds. Conventional methods for the structural one of bioactive compounds are usually time-consuming and labor-intensive procedures that include the isolation of individual compounds by a combination of chromatographic techniques and structure elucidation through a combination of different methods [20]. The final structure confirmation of a new compound is always performed with a set of independent methods, such as nuclear magnetic resonance (NMR) spectroscopy, mass spectrometry (MS), or other analytical methods and chemical transformation. Despite the instrumentation developments of recent years, the analysis of bioactive natural compounds using conventional approaches remains a challenging task due to the difficulty of isolating individual compounds from fractions consisting of many components and with high chemical diversity covering a broad concentration range. As a result, only certain compounds can be described; overall, the entire metabolite pool remains poorly studied.
At present, modern mass spectrometry techniques are widely employed for the identification and structural analysis of novel natural compounds [34][35][36]. Hyphenated techniques combining various separation methods with mass spectrometry are applied for metabolomic and target profiling and allow for the characterization of compounds in complex mixtures extracted from biological material. Different MS-imaging techniques precisely localize and quantify the metabolites in tissues [37]. Recently developed ion mobility (IM) methods add dimension to conventional chromatography separation, allowing the stereoisomers that cannot be separated by liquid chromatography (LC) to be identified [38].
Mass spectrometry is used in various fields of marine sciences today, including marine proteomics [39], metabolomics [40][41], lipidomics [42], marine toxicology [43], ecology studies [44], and others. The introduction of modern mass-spectrometric approaches has greatly contributed to the development of metabolomics as a transdisciplinary science that aims at the qualitative and quantitative determination of the whole metabolite pool of organisms. Although no single analytical method exists that can determine all members of the metabolome simultaneously, MS-based metabolomics has been successfully applied to analyze a wide range of compounds from various sources. In recent years, MS-based metabolomics has emerged as a useful tool in natural product research. In addition to metabolic fingerprinting, two approaches used in metabolomics studies can be distinguished [45]. The first, metabolic profiling, focuses on the analysis of structure-related metabolites or metabolites related to a specific metabolic pathway. Such an approach provides information on the chemical composition of extracts or fractions and allows for the dereplication of known bioactive compounds and detection of new compounds as well as the evaluation of the their isolation possibility [34]. The metabolome-oriented approach aims to detect differences in metabolic profiles that occur in response to stress, disease, changing environmental conditions, or other influences in comparative experiments.
Metabolomic studies of marine organisms is a field that uses a variety of modern approaches, including MS, NMR methods, and hyphenated techniques [40][41][44].
2. Overview of MS-Based Metabolomic Workflows in the Analysis of Starfish and Sea Cucumber Bioactive Compounds
In terms of workflow, a typical MS-based metabolomic one involves the stages of sample collection, extraction, fractionation and/or purification, measurement, identification, and analysis of the results (Figure 1). The analytical protocols used in MS-based marine metabolomics have several important differences from those used to analyze the metabolites of terrestrial animals and plants [41]. This section discusses the characteristic features of MS-based metabolomic approaches in the context of starfish and sea cucumber bioactive compounds, including steps of sample preparation, acquisition, and analysis.
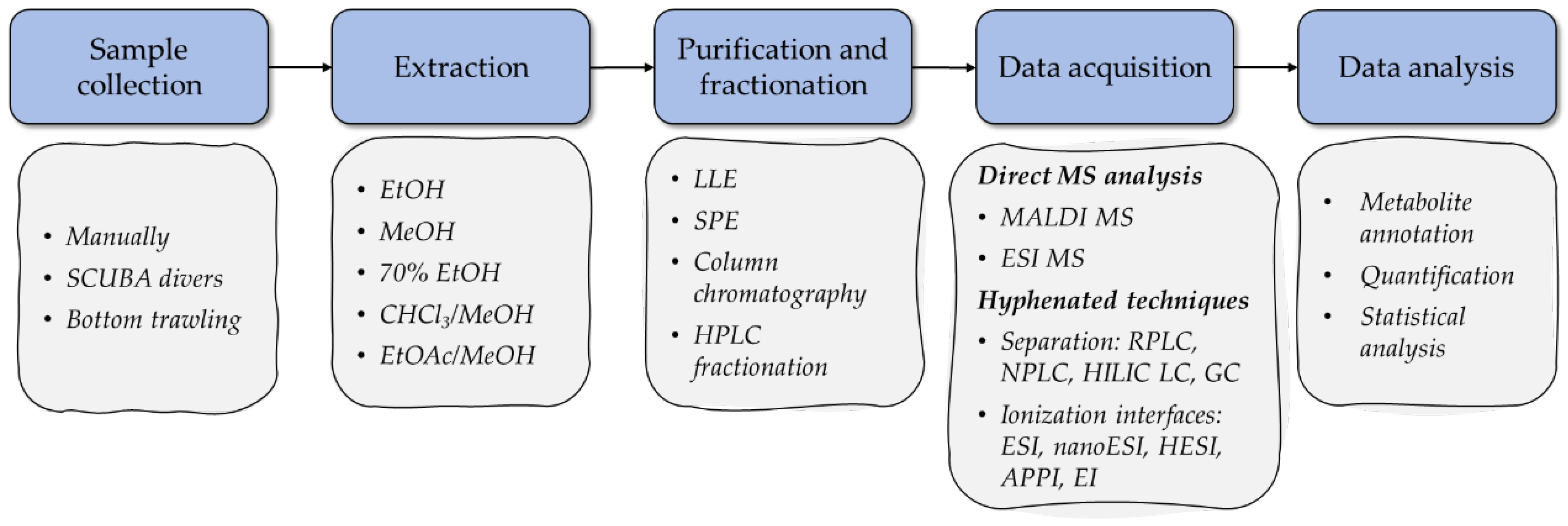
Figure 1. Main research stages in MS-based metabolomics studies of starfish and sea cucumber bioactive compounds.
2.1. Sample Preparation
The sample preparation stage, which comprises sample collection and extraction, is the most important in metabolomics research. Most of the studied starfish and sea cucumbers are collected from the coastal area manually or by SCUBA divers, or, if the depth exceeds 30 m, by bottom trawling. The main difficulties encountered in the collection of echinoderms are related to accessibility and their limited quantity. Many starfish and sea cucumbers are common species found in coastal areas where collection is unproblematic, while others occur in restricted or inaccessible geographic areas or in limited populations. Certain sea cucumber species, such as Apostichopus japonicus and Holothuria scabra, are aquaculture species, making their collection much simpler than the collection of wild specimens. In contrast to terrestrial ecosystems, when collecting marine samples the depth, salinity, and oxygen concentration of the water must be considered in addition to general factors such as temperature and light. A specimen’s location, physiological state, sex, and season can have a great metabolic influence. Difficulties are often caused by significant distances between the collection site and the laboratory, which requires more complicated logistics and specific sample preparation protocols.
Stress caused by handling induces responses in animals at the biochemical level [46] and can cause sea cucumber evisceration, the expulsion of the internal organs from the body [47]. To avoid such changes, as well as metabolomic changes resulting from enzymatic turnover during transportation or sample processing, it is highly recommended to quench the metabolism rapidly [48]. There are protocols designed for quenching, including flash-freezing using liquid nitrogen or dry ice, lyophilization, and freeze-drying; however, some of these are difficult to implement when animals are collected in the wild. Therefore, in most cases researchers use alternative protocols such as freezing or direct extraction with organic solvents [41].
The collected sample material must be processed to extract the metabolites of interest and remove salts and impurities. Extractions with organic solvents are commonly used for this purpose. Due to the high chemical diversity of metabolites, there is no single solvent capable of capturing all the required compounds without related impurities and contaminants. Generally, polar and semi-polar metabolites such as triterpene glycosides, asterosaponins, and gangliosides are preferentially extracted with hydro-alcoholic solutions, while lipid, sterol, terpene, and other non-polar compound extraction can be achieved with hydrophobic solvents (chloroform, hexane) or liquid–liquid extraction (LLE) by Folch’s [49] and Bligh and Dyer’s [50] methods. Extraction with methyl tert-butyl ether (MTBE) [51] can be used for the recovery of both polar and non-polar metabolites into separate fractions. In addition, the selected extraction protocols and solvents must be related to the analytical methods used.
It should be noted that most starfish and sea cucumber extracts contain significant amounts of salts, even if a non-polar solvent is used for extraction. Such samples are incompatible with analytical techniques such as mass spectrometry and NMR because of the effect of salts on analytical performance. For example, the presence of a small concentration of NaCl can cause the appearance of unexpected adducts at ESI MS, while larger concentrations can suppress analyte ionization and lead to salt crystal deposits in the ion source and the capillary, which can cause the instrument malfunction. MALDI MS is more tolerant to salt impurities and can be used for preliminary screening of extracts without additional purification. Desalting of marine extracts typically involves column chromatography (CC), liquid–liquid extraction, and solid-phase extraction (SPE).
Another problem can be the presence of lipid impurities and/or proteins in samples of polar secondary metabolites. For example, when extracting starfish polar steroid compounds or sea cucumber triterpene glycosides the crude hydro-alcoholic extracts may contain a large concentration of phospholipids, which can complicate chromatographic separations and suppress the ionization of the target analytes. If lipid compounds are not included in the target pool, additional purification of the extract can improve both LC separation and MS identification of target analytes. In order to remove such interfering compounds, column chromatography with Amberlite XAD-4, Sephadex LH-60, or other sorbents, LLE or SPE is usually used. In order to simplify the analysis of extremely complex extracts and obtain a mixture containing only structure-related metabolites of interest, fractionation using column chromatography, flash chromatography, or HPLC is used.
The choice of extraction solvent and purification methods affects the efficiency of the sample preparation stage. The use of unsuitable solvents and extraction methods can result in quantitatively and qualitatively incomplete extraction, while the use of suboptimal purification or fractionation procedures can lead to loss of the target metabolites. To the best, there are no published studies comparing the effectiveness of the most commonly used sample preparation protocols for the analysis of starfish and sea cucumber bioactive compounds.
2.2. Data Acquisition
Structural elucidation of starfish and sea cucumber bioactive compounds remains a difficult task due to the great diversity of these compounds and the complexity of the analyzed mixtures. Usually, these compounds form very complicated mixtures which are difficult to separate into pure compounds by chromatography. In the past, the application of chemical methods was required in order to identify the structure of such compounds. In particular, acid hydrolysis was used to recognize steroid and triterpene glycoside structures. While this approach allowed the partial characterization of aglycon structures and the determination of qualitative and quantitative monosaccharide composition, the destruction of native aglycon was frequent.
For a long time, electron ionization (EI) was the only possible mass spectrometry technique. Rashkes et al. carried out mass spectrometry research on six polyhydroxysteroid compounds and glycosides isolated from the Far Eastern starfish Patiria pectinifera and determined the characteristic fragmentation pattern of starfish polyhydroxysteroid under EI conditions [52]. EI and GC-EI MS were widely used for the determination of structures of aglycones and oligosaccharide chains of asterosaponins and triterpene glycosides after hydrolysis of glycosides and chemical derivatization of monosaccharides [20]. GC coupled with EI MS remains one of the most suitable metabolomic techniques for analyzing the wide range of volatile, semi-volatile non-polar compounds and derivatized polar metabolites. Electron impact ionization results in highly reproducible fragmentation patterns that can be used for identification by database search along with retention times indexes.
The application of fast atom bombardment (FAB) MS allows for analysis of the more polar and unstable compounds. Introduced in 1983, FAB has been successfully used for the determination of the structures of starfish steroid glycosides and sea cucumber triterpene glycosides as well as cerebrosides and gangliosides, which could not be analyzed by EI MS [53]. FAB mass spectra of starfish and sea cucumber glycosides can show molecular ions as well as fragmentation products, providing information about molecular formulae, the presence and location of sulfate groups, the structures of carbohydrate chains, and aglycon. Collision-induced dissociation (CID) experiments can provide additional structural information on the structural features of aglycon, the quantity and type of monosaccharides attached to aglycon, and their location.
Electrospray ionization (ESI) and Matrix-Assisted Laser Desorption/Ionization (MALDI) have significantly expanded the possibilities of mass spectrometry for the analysis of natural products. ESI has had an enormous impact on the analysis of polar and non-volatile molecules as well as large biomolecules. In contrast to electron ionization, in-source fragmentation under ESI conditions is practically unrealized; tandem MS methods are used to initiate the fragmentation of these ions. ESI mass spectrometry is currently the most common ionization technique; it has been widely used for the characterization of natural compounds, including steroid and triterpene glycosides, polar lipids, and other compounds from purified starfish and sea cucumber extracts. MALDI MS is another efficient method for the analysis of natural compounds. The necessity of using matrices and the presence of matrix ion peaks at spectra in the low mass range are drawbacks; however, due to its high sensitivity, high speed of analysis, and tolerance to inorganic salts impurities, MALDI MS is widely used for rapid screening and chemical characterization of complex mixtures. Recent advances in analytical techniques, including high-resolution time-of-flight (TOF), Fourier transform (FT), and Orbitrap mass analyzers have high scan speeds along with extended dynamic range and sensitivity, allowing for the development of hybrid instruments and new ionization interfaces such as nanoelectrospray (nanoESI) and heated electrospray ionization (HESI) and leading to the establishment of high-throughput protocols for the analysis of the most complex mixtures of natural compounds. The development of hyphenated techniques combining liquid chromatography or gas chromatography with mass spectrometry (LC-MS or GC-MS) makes allows for straightforward analysis of the compounds present in complicated extracts.
2.3. Data Analysis
Data analysis is the next important stage of MS-based research. Generally, the processing of data obtained using chromatography-MS methods has included the steps of identifying m/z signals, chromatographic peak detection, filtering, alignment, and identification [54]. Many freely available (XCMS [55], MZmine 2 [56], OpenMS [57], and MS-DIAL [58]) and commercial software tools are currently available for the processing of LC-MS and GC-MS data. While the processing of GC-MS data is well-established and relatively simple, the results are usually limited to known compounds presented in databases. LC-MS is a more versatile method, covering broad chemistries and sensitivity ranges, although it produces more complex data. Due to the lower resolution and reproducibility of LC separation and the presence of adduct, isotope, fragment, and contamination peaks in ESI spectra, LC-MS data processing is much more difficult.
In certain cases, special approaches are useful for data processing. In order to process MS profiling data, methods based on scanning neutral losses, characteristic fragments, and an in-house library for rapid screening of the compounds of interest are often used. For example, the construction of ion chromatograms for negative fragment ions at m/z 96.96 can be used for detecting sulfated compounds like asterosaponins and sulfated triterpene glycosides, and cerebrosides can be detected according to the neutral loss fragments of 180 Da [59].
Similar to common metabolomic studies, metabolite identification is a current bottleneck in the analysis of starfish and sea cucumber metabolites. Chromatography-MS-based analysis can result in a huge number of peaks that are extremely difficult to identify. Even when analyzing well-studied organisms, only small percentages of the data collected in a typical LC-MS experiment can be matched to known molecules [60]. The chemical composition of starfish and sea cucumbers remains poorly investigated and the percentage of identified compounds can be extremely low. According to Metabolomics Standards Initiative recommendations, high identification confidence can be obtained by comparing an accurate high-resolution monoisotopic mass, MS/MS spectra, and retention times with data from an authentic chemical standard [61]. However, the available libraries of certified standards do not cover the entire scope of biochemical diversity, especially in the area of marine bioactive compounds. Although only putative annotation is possible without matching experimental data to data for authentic chemical standards [61], the availability of comprehensive open-access databases is extremely important for the successful application of mass spectrometry to the analysis of complex mixtures of natural compounds. Existing databases cover various natural compounds [62], and several databases, such as the GNPS database [63], MassBank [64], Metlin [65], the Human Metabolome Database [66], and MassBank of North America (https://mona.fiehnlab.ucdavis.edu, accessed on 1 April 2022) include MS/MS spectra of natural compounds from different sources. Databases such as the Dictionary of Marine Natural Products (https://dmnp.chemnetbase.com, accessed on 1 April 2022) and MarinLit (https://marinlit.rsc.org, accessed on 1 April 2022) include structural information, and the NMR and UV spectra of marine-derived compounds, although MS and MS/MS data on marine natural compounds in all existing databases is extremely limited. There are currently no databases covering taxonomic, structural, and experimental mass spectrometry data on bioactive metabolites of marine echinoderms.
Moreover, unlike peptides, oligosaccharides, and lipids, the MS fragmentation of most secondary metabolites is less studied due to the vast structural variability, and the de novo identification of metabolites by MS/MS spectra is very difficult. Several computational approaches based on machine learning or quantum chemistry calculations have been proposed for the in silico generation of MS/MS spectra or the prediction of structural features of compounds based on the experimental MS/MS spectra [67][68]. The molecular networking approach is based on the clustering of detected compounds by the similarity of their MS/MS spectra and allows for the annotation of related metabolites [63]. Using models for the in silico prediction of LC retention times can help to improve the reliability of identification in metabolomics analysis [69]. However, despite the recent advances in computational approaches, the currently used algorithms need to be significantly improved before effective identification and structural elucidation of marine bioactive compounds is possible. Along with the huge degree of structural variability, these issues limit the application of MS for annotation, dereplication, and structural elucidation in studies of metabolites from marine organisms.
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173.
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2020, 37, 175–223.
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2021, 23, 9–10.
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2022.
- Jiménez, C. Marine natural products in medicinal chemistry. ACS Med. Chem. Lett. 2018, 9, 959–961.
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53.
- Zhang, L.-X.; Fan, X.; Shi, J.-G. A novel pyrrole oligoglycoside from the starfish Asterina pectinifera. Nat. Prod. Res. 2006, 20, 229–233.
- Yang, X.W.; Chen, X.Q.; Dong, G.; Zhou, X.F.; Chai, X.Y.; Li, Y.Q.; Yang, B.; Zhang, W.D.; Liu, Y. Isolation and structural characterisation of five new and 14 known metabolites from the commercial starfish Archaster typicus. Food Chem. 2011, 124, 1634–1638.
- Purcell, S.W. Managing sea cucumber fisheries with an ecosystem approach. In FAO Fisheries and Aquaculture Technical Paper; No. 520; Lovatelli, A., Vasconcellos, M., Yimin, Y., Eds.; FAO: Rome, Italy, 2010; 157p, ISBN 978-92-5-106489-4.
- Bordbar, S.; Anwar, F.; Saari, N. High-value components and bioactives from sea cucumbers for functional foods—A review. Mar. Drugs 2011, 9, 1761–1805.
- Li, Y.X.; Himaya, S.W.A.; Kim, S.K. Triterpenoids of marine origin as anti-cancer agents. Molecules 2013, 18, 7886–7909.
- Gomes, A.R.; Freitas, A.C.; Rocha-Santos, T.A.P.; Duarte, A.C. Bioactive compounds derived from echinoderms. RSC Adv. 2014, 4, 29365–29382.
- Khotimchenko, Y. Pharmacological potential of sea cucumbers. Int. J. Mol. Sci. 2018, 19, 1342.
- Lazzara, V.; Arizza, V.; Luparello, C.; Mauro, M.; Vazzana, M. Bright spots in the darkness of cancer: A review of starfishes-derived compounds and their anti-tumor action. Mar. Drugs 2019, 17, 617.
- Zhou, Y.; Farooqi, A.A.; Xu, B. Comprehensive review on signaling pathways of dietary saponins in cancer cells suppression. Crit. Rev. Food Sci. Nutr. 2021, 9, 1–26.
- Wen, M.; Cui, J.; Xu, J.; Xue, Y.; Wang, J.; Xue, C.; Wang, Y. Effects of dietary sea cucumber saponin on the gene expression rhythm involved in circadian clock and lipid metabolism in mice during nighttime-feeding. J. Physiol. Biochem. 2014, 70, 801–808.
- Chumphoochai, K.; Chalorak, P.; Suphamungmee, W.; Sobhon, P.; Meemon, K. Saponin-enriched extracts from body wall and Cuvierian tubule of Holothuria leucospilota reduce fat accumulation and suppress lipogenesis in Caenorhabditis elegans. J. Sci. Food Agric. 2019, 99, 4158–4166.
- Liu, X.; Xu, J.; Xue, Y.; Gao, Z.; Li, Z.; Leng, K.; Wang, J.; Xue, C.; Wang, Y. Sea cucumber cerebrosides and long-chain bases from Acaudina molpadioides protect against high fat diet-induced metabolic disorders in mice. Food Funct. 2015, 6, 3428–3436.
- Luparello, C.; Mauro, M.; Lazzara, V.; Vazzana, M. Collective locomotion of human cells, wound healing and their control by extracts and isolated compounds from marine invertebrates. Molecules 2020, 25, 2471.
- Minale, L.; Riccio, R.; Zollo, F. Steroidal Oligoglycosides and polyhydroxysteroids from echinoderms. Prog. Chem. Org. Nat. Prod. 1993, 62, 75–308.
- Chludil, H.D.; Murray, A.P.; Seldes, A.M.; Maier, M.S. Biologically active triterpene glycosides from sea cucumbers (Holothuroidea, Echinodermata). In Studies in Natural Products Chemistry; Atta-ur-Rahman, Ed.; Elsevier Science Publisher: Amsterdam, The Netherlands, 2003; Volume 28, pp. 587–615.
- Kim, S.; Himaya, S.W.A. Triterpene glycosides from sea cucumbers and their biological activities. Adv. Food Nutr. Res. 2012, 65, 297–319.
- Kalinin, V.I.; Silchenko, A.S.; Avilov, S.A.; Stonik, V.A. Progress in the studies of triterpene glycosides from sea cucumbers (Holothuroidea, Echinodermata) Between 2017 and 2021. Nat. Prod. Commun. 2021, 16, 1934578X211053934.
- Kalinin, V.I.; Avilov, S.A.; Silchenko, A.S.; Stonik, V.A.; Elyakov, G.B. Triterpene glycosides of sea cucumbers (Holothuroidea, Echinodermata) as taxonomic markers. Nat. Prod. Commun. 2015, 10, 21–26.
- Kalinin, V.I.; Silchenko, A.S.; Avilov, S.A.; Stonik, V.A. Non-holostane aglycones of sea cucumber triterpene glycosides. Structure, biosynthesis, evolution. Steroids 2019, 147, 42–51.
- Stonik, V.A.; Ivanchina, N.V.; Kicha, A.A. New polar steroids from starfish. Nat. Prod. Commun. 2008, 3, 1587–1610.
- Ivanchina, N.V.; Kicha, A.A.; Stonik, V.A. Steroid glycosides from marine organisms. Steroids 2011, 76, 425–454.
- Dong, G.; Xu, T.; Yang, B.; Lin, X.; Zhou, X.; Yang, X.; Liu, Y. Chemical constituents and bioactivities of starfish. Chem. Biodivers. 2011, 8, 740–791.
- Ivanchina, N.; Kicha, A.; Malyarenko, T.; Stonik, V. Recent studies of polar steroids from starfish: Structures, biological activities and biosynthesis. In Advances in Natural Products Discovery; Nova Science Publishers: New York, NY, USA, 2017; pp. 191–224.
- Xia, J.M.; Miao, Z.; Xie, C.L.; Zhang, J.W.; Yang, X.W. Chemical constituents and bioactivities of starfishes: An update. Chem. Biodivers. 2020, 17, e1900638.
- Stonik, V.A.; Kicha, A.A.; Malyarenko, T.V.; Ivanchina, N.V. Asterosaponins: Structures, taxonomic distribution, biogenesis and biological activities. Mar. Drugs 2020, 18, 584.
- Kalinin, V.I.; Aminin, D.L.; Avilov, S.A.; Silchenko, A.S.; Stonik, V.A. Triterpene glycosides from sea cucucmbers (Holothurioidea, Echinodermata). Biological activities and functions. In Studies in Natural Products Chemistry (Bioactive Natural Products); Atta-ur-Rahman, Ed.; Elsevier Science Publisher: Amsterdam, The Netherlands, 2008; Volume 35, pp. 135–196. ISBN 1572-5995.
- Careaga, V.P.; Maier, M.S. Cytotoxic triterpene glycosides from sea cucumbers. In Handbook of Anticancer Drugs from Marine Origin; Kim, S.-K., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 515–528. ISBN 978-3-319-07145-9.
- Wolfender, J.L.; Litaudon, M.; Touboul, D.; Queiroz, E.F. Innovative omics-based approaches for prioritisation and targeted isolation of natural products-new strategies for drug discovery. Nat. Prod. Rep. 2019, 36, 855–868.
- Alvarez-Rivera, G.; Ballesteros-Vivas, D.; Parada-Alfonso, F.; Ibañez, E.; Cifuentes, A. Recent applications of high resolution mass spectrometry for the characterization of plant natural products. TrAC Trends Anal. Chem. 2019, 112, 87–101.
- Aydoğan, C. Recent advances and applications in LC-HRMS for food and plant natural products: A critical review. Anal. Bioanal. Chem. 2020, 412, 1973–1991.
- Boughton, B.A.; Thinagaran, D.; Sarabia, D.; Bacic, A.; Roessner, U. Mass spectrometry imaging for plant biology: A review. Phytochem. Rev. 2016, 15, 445–488.
- Zhang, X.; Quinn, K.; Cruickshank-Quinn, C.; Reisdorph, R.; Reisdorph, N. The application of ion mobility mass spectrometry to metabolomics. Curr. Opin. Chem. Biol. 2018, 42, 60–66.
- Chandramouli, K. Marine proteomics: Challenges and opportunities. J. Data Min. Genom. Proteom. 2016, 7, 3–4.
- Stuart, K.A.; Welsh, K.; Walker, M.C.; Edrada-Ebel, R.A. Metabolomic tools used in marine natural product drug discovery. Expert Opin. Drug Discov. 2020, 15, 499–522.
- Bayona, L.M.; de Voogd, N.J.; Choi, Y.H. Metabolomics on the study of marine organisms. Metabolomics 2022, 18, 17.
- Yeo, J.D.; Parrish, C.C. Mass spectrometry-based lipidomics in the characterization of individual triacylglycerol (TAG) and phospholipid (PL) species from marine sources and their beneficial health effects. Rev. Fish. Sci. Aquac. 2022, 30, 81–100.
- Christian, B.; Luckas, B. Determination of marine biotoxins relevant for regulations: From the mouse bioassay to coupled LC-MS methods. Anal. Bioanal. Chem. 2008, 391, 117–134.
- Goulitquer, S.; Potin, P.; Tonon, T. Mass spectrometry-based metabolomics to elucidate functions in marine organisms and ecosystems. Mar. Drugs 2012, 10, 849.
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171.
- Tonn, N.; Novais, S.C.; Silva, C.S.E.; Morais, H.A.; Correia, J.P.S.; Lemos, M.F.L. Stress responses of the sea cucumber Holothuria forskali during aquaculture handling and transportation. Mar. Biol. Res. 2016, 12, 948–957.
- Ding, K.; Zhang, L.; Huo, D.; Guo, X.; Liu, X.; Zhang, S. Metabolomic analysis of coelomic fluids reveals the physiological mechanisms underlying evisceration behavior in the sea cucumber Apostichopus Jpn. Aquac. 2021, 543, 736960.
- Vuckovic, D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2012, 403, 1523–1548.
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509.
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917.
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146.
- Rashkes, Y.V.; Kicha, A.A.; Levina, E.V.; Stonik, V.A. Mass spectra of polyhydrosysteroids of the starfish Patiria pectinifera. Chem. Nat. Compd. 1985, 21, 337–342.
- Komori, T.; Nanri, H.; Itakura, Y.; Sakamoto, K.; Taguchi, S.; Higuchi, R.; Kawasaki, T.; Higuchi, T. Biologically active glycosides from Asteroidea, III. Steroid oligoglycosides from the starfish Acanthaster planci L., 2. Structures of two newly characterized genuine sapogenins and an oligoglycoside sulfate. Liebigs Ann. Der Chem. 1983, 1983, 37–55.
- Alonso, A.; Marsal, S.; JuliÃ, A. Analytical methods in untargeted metabolomics: State of the art in 2015. Front. Bioeng. Biotechnol. 2015, 3, 23.
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787.
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395.
- Röst, H.L.; Sachsenberg, T.; Aiche, S.; Bielow, C.; Weisser, H.; Aicheler, F.; Andreotti, S.; Ehrlich, H.C.; Gutenbrunner, P.; Kenar, E.; et al. OpenMS: A flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 2016, 13, 741–748.
- Tsugawa, H.; Ikeda, K.; Takahashi, M.; Satoh, A.; Mori, Y.; Uchino, H.; Okahashi, N.; Yamada, Y.; Tada, I.; Bonini, P.; et al. A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 2020, 38, 1159–1163.
- Jia, Z.; Cong, P.; Zhang, H.; Song, Y.; Li, Z.; Xu, J.; Xue, C. Reversed-phase liquid chromatography–quadrupole-time-of-flight mass spectrometry for high-throughput molecular profiling of sea cucumber cerebrosides. Lipids 2015, 50, 667–679.
- Aksenov, A.A.; Da Silva, R.; Knight, R.; Lopes, N.P.; Dorrestein, P.C. Global chemical analysis of biology by mass spectrometry. Nat. Rev. Chem. 2017, 1, 0054.
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221.
- Wolfender, J.L.; Nuzillard, J.M.; Van Der Hooft, J.J.J.; Renault, J.H.; Bertrand, S. Accelerating metabolite identification in natural product research: Toward an ideal combination of liquid chromatography-high-resolution tandem mass spectrometry and nmr profiling, in silico databases, and chemometrics. Anal. Chem. 2019, 91, 704–742.
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837.
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714.
- Smith, C.A.; Maille, G.O.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN a metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751.
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617.
- Krettler, C.A.; Thallinger, G.G. A map of mass spectrometry-based in silico fragmentation prediction and compound identification in metabolomics. Brief. Bioinform. 2021, 22, bbab073.
- Böcker, S. Searching molecular structure databases using tandem MS data: Are we there yet? Curr. Opin. Chem. Biol. 2017, 36, 1–6.
- Stanstrup, J.; Neumann, S.; Vrhovšek, U. PredRet: Prediction of retention time by direct mapping between multiple chromatographic systems. Anal. Chem. 2015, 87, 9421–9428.
More
Information
Subjects:
Biochemical Research Methods
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.0K
Revisions:
2 times
(View History)
Update Date:
20 May 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




