
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Johane Hans Bracamonte | -- | 5628 | 2022-05-18 01:00:18 | | | |
| 2 | Lindsay Dong | -23 word(s) | 5605 | 2022-05-18 04:30:42 | | |
Video Upload Options
Inverse modeling approaches in cardiovascular medicine are a collection of methodologies that can provide non-invasive patient-specific estimations of tissue properties, mechanical loads, and other mechanics-based risk factors using medical imaging as inputs. Its incorporation into clinical practice has the potential to improve diagnosis and treatment planning with low associated risks and costs. Inverse method applications are multidisciplinary, requiring tailored solutions to the available clinical data, pathology of interest, and available computational resources.
1. Introduction
2. Governing Principles of Biomechanics
2.1. Structural Mechanics of Cardiovascular Tissue
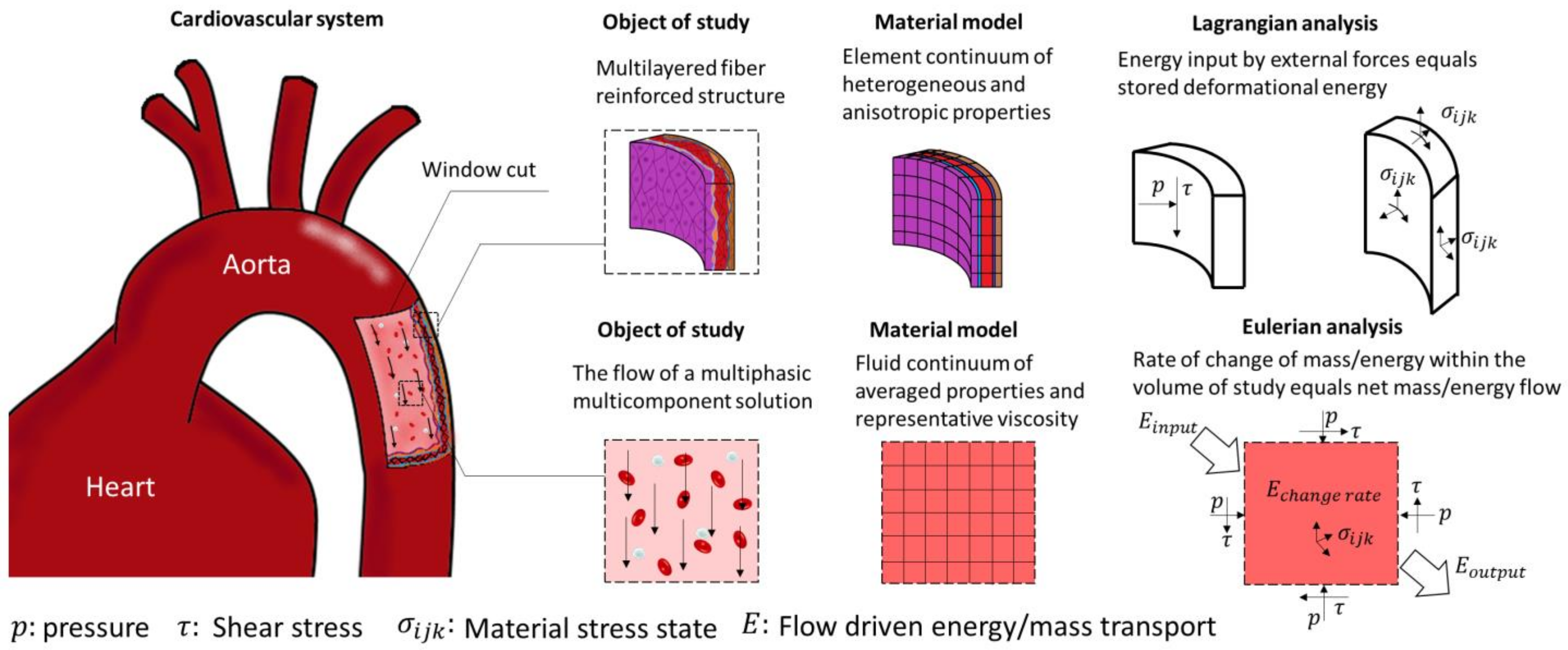
2.2. Fluid Mechanics of Blood Flow
2.3. Fluid-Structure Interactions (FSI)
3. Numerical Methods
3.1. Finite Volume Method
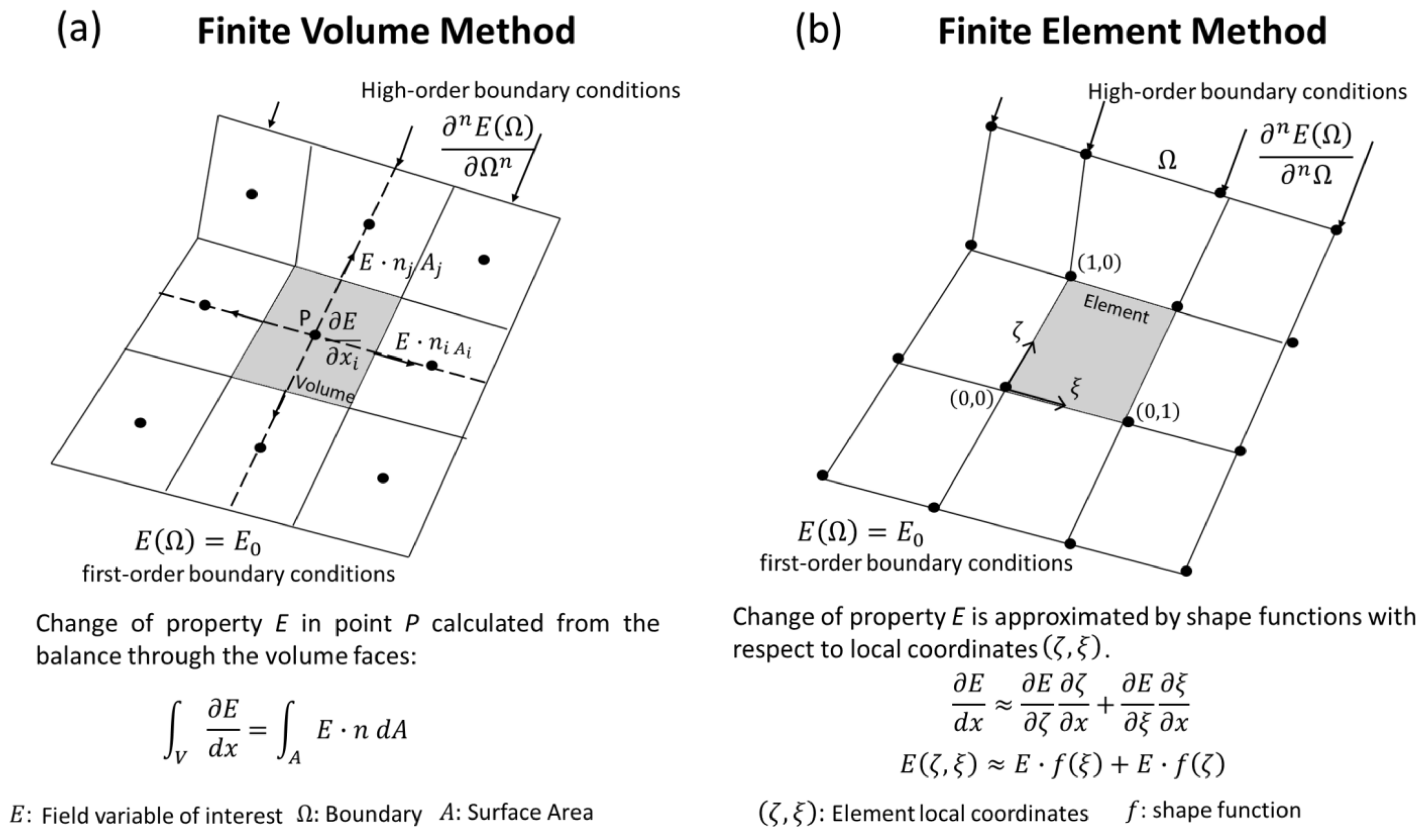
3.2. Finite Element Method (FEM)
4. Inverse Problems
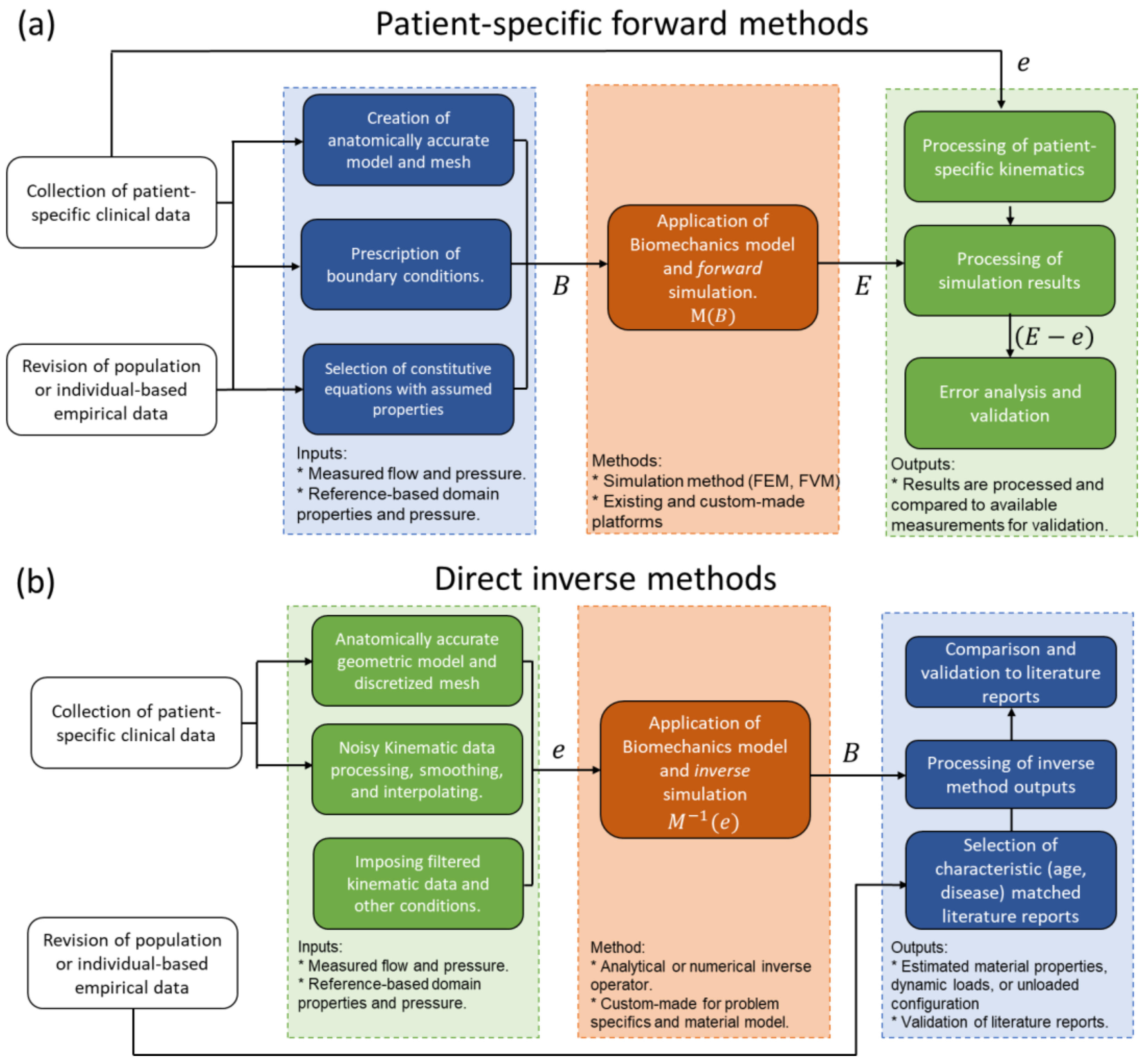
4.1. Direct Inverse Methods
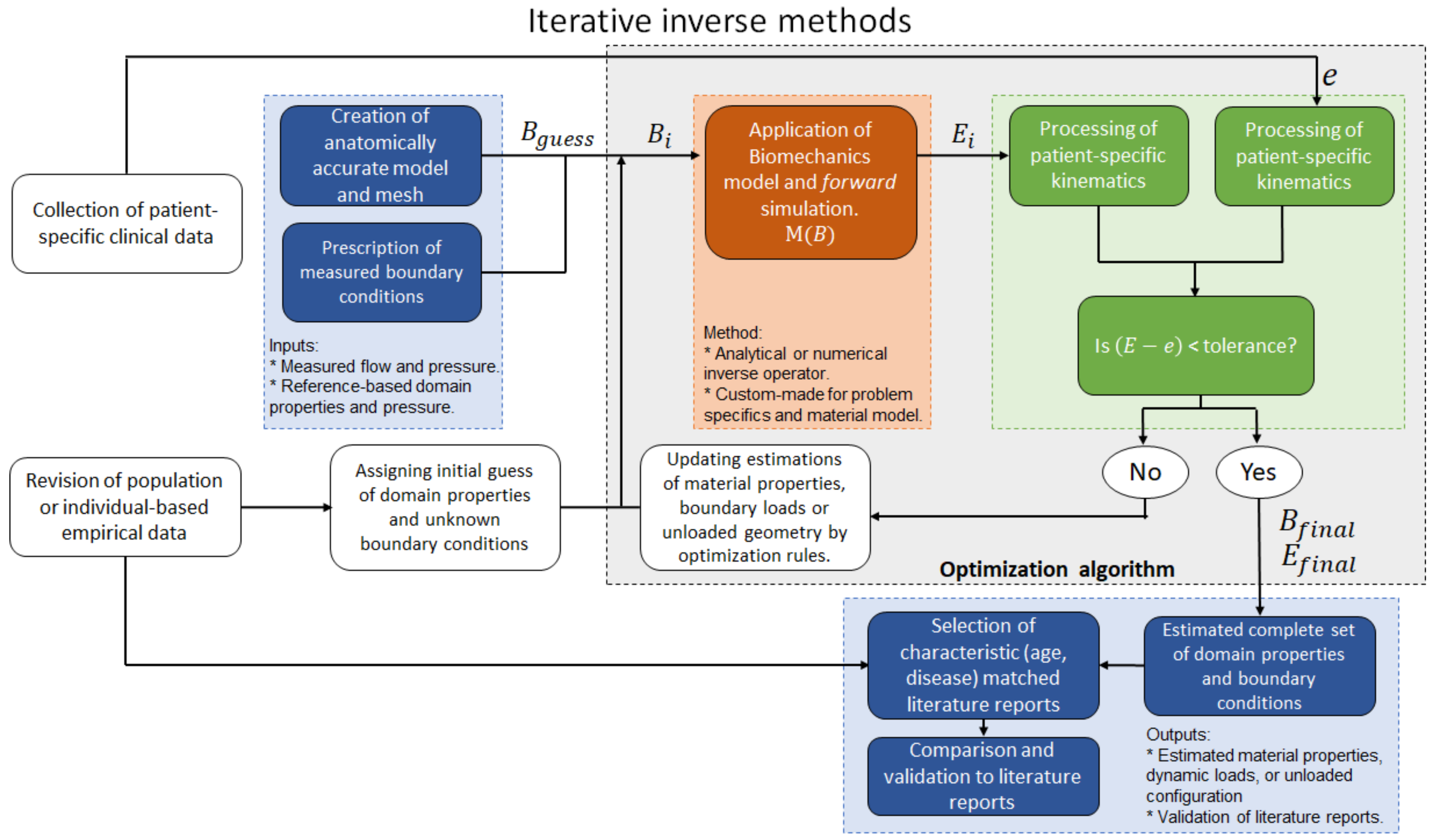
4.2. Iterative Inverse Methods
5. Medical Imaging-Based Kinematics
5.1. Ultrasound Technology (US)
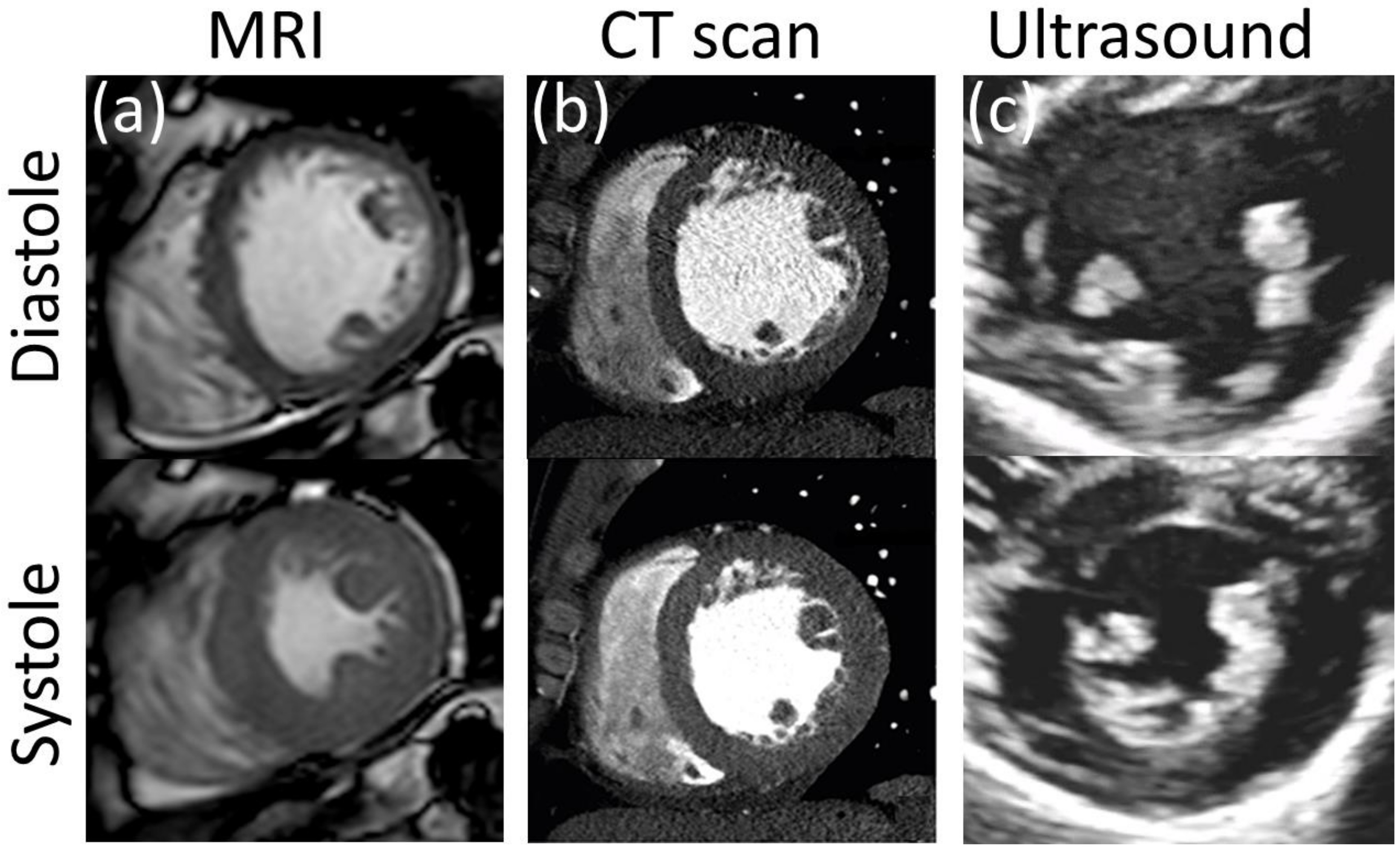
5.2. Magnetic Resonance Imaging (MRI)
There are several options for MRI-based assessment of kinematics of both cardiovascular soft tissues and blood flow. Four prominent examples are: MRI tagging and displacement encoding with stimulated echoes (DENSE) for tissue displacement, and phase contrast and 4D flow MRI for blood flow quantification.
5.3. Computerized Tomography (CT)
6. Applications to Cardiovascular Medicine
6.1. The Unloaded Reference Configuration in Cardiovascular Mechanics
6.2. The Heart
6.3. Valves and Leaflets
Leaflets are typically thin structures showing complex displacement patterns, which renders them extremely challenging to resolve through in vivo imaging techniques. In vivo inverse modeling of ovine heart valves function has been achieved by the use of fluoroscopic markers implanted on the surface of mitral valve leaflets [95][96], a technique that cannot be pursued in human studies. More recently, Lee et al. applied ultrasound technology to assess the anatomy and displacement of the mitral valve of ovine animal models to explore the use of inverse modeling, and in vivo mechanical properties and stress distribution were successfully estimated [97][98]. Aggarwal et al. estimated the residual strain on human aortic valves by combining in vivo imaging with measurements on explanted tissues [99]. The authors collected in vivo transesophageal 3D echocardiographic images of the aortic valve from five open-heart transplant patients at three configurations: fully open, just-coapted, and fully loaded.
6.4. Arterial Wall
6.5. Hemodynamics
Zambrano et al. proposed an iterative inverse method for the study of the pulmonary artery [113]. Intravascular pressure measurements, PC MRI at the main branches of the pulmonary artery, and cine MRI were collected from a pulmonary hypertensive adult patient and a healthy volunteer with no reported cardiovascular disease. A 3D model from the main pulmonary artery (MPA) down to the 4th branch generation was built from MRI images at the end-diastole configuration, which was considered stress-free. MRI-derived diameter changes were calculated at the main pulmonary artery and coupled to pressure measurements.The proposed model was able to reproduce the expected increase in arterial stiffness and vascular flow resistance in the hypertensive patient. In a follow-up study, the methodology was applied to a cohort of six individuals with pulmonary artery hypertension and five healthy volunteers [114].
References
- Kakisis, J.D.; Liapis, C.D.; Sumpio, B.E. Effects of Cyclic Strain on Vascular Cells. Endothelium 2004, 11, 17–28.
- Chang, H.-I.; Wang, Y. Cell response to surface and architecture of tissue engineering scaffolds. In Regenerative Medicine and Tissue Engineering—Cells and Biomaterials; Eberli, D., Ed.; InTech Open Access Publisher: Rijeka, Croatia, 2011.
- Butler, D.L.; Goldstein, S.A.; Guilak, F. Functional Tissue Engineering: The Role of Biomechanics. J. Biomech. Eng. 2000, 122, 570–575.
- Tang, B.T.; Pickard, S.S.; Chan, F.P.; Tsao, P.S.; Taylor, C.A.; Feinstein, J.A. Wall Shear Stress is Decreased in the Pulmonary Arteries of Patients with Pulmonary Arterial Hypertension: An Image-Based, Computational Fluid Dynamics Study. Pulm. Circ. 2012, 2, 470–476.
- Pourmodheji, R.; Jiang, Z.; Tossas-Betancourt, C.; Figueroa, C.A.; Baek, S.; Lee, L.-C. Inverse modeling framework for characterizing patient-specific microstructural changes in the pulmonary arteries. J. Mech. Behav. Biomed. Mater. 2021, 119, 104448.
- Watton, P.N.; Hill, N.A. Evolving mechanical properties of a model of abdominal aortic aneurysm. Biomech. Model. Mechanobiol. 2009, 8, 25–42.
- Neal, M.L.; Kerckhoffs, R. Current progress in patient-specific modeling. Brief. Bioinform. 2010, 11, 111–126.
- Marsden, A.; Esmaily-Moghadam, M. Multiscale Modeling of Cardiovascular Flows for Clinical Decision Support. Appl. Mech. Rev. 2015, 67, 030804.
- Itatani, K.; Miyazaki, S.; Furusawa, T.; Numata, S.; Yamazaki, S.; Morimoto, K.; Makino, R.; Morichi, H.; Nishino, T.; Yaku, H. New imaging tools in cardiovascular medicine: Computational fluid dynamics and 4D flow MRI. Gen. Thorac. Cardiovasc. Surg. 2017, 65, 611–621.
- Wilson, J.S.; Zhong, X.; Hair, J.B.; Taylor, W.R.; Oshinski, J.N. In Vivo Quantification of Regional Circumferential Green Strain in the Thoracic and Abdominal Aorta by Two-Dimensional Spiral Cine DENSE MRI. J. Biomech. Eng. 2019, 141, 0609011–06090111.
- Kong, F.; Shadden, S.C. Automating Model Generation for Image-Based Cardiac Flow Simulation. J. Biomech. Eng. 2020, 142, 1110111–11101113.
- Markl, M.; Schnell, S.; Barker, A. 4D Flow Imaging: Current Status to Future Clinical Applications. Curr. Cardiol. Rep. 2014, 16, 481.
- Wilson, J.S.; Taylor, W.R.; Oshinski, J. Assessment of the regional distribution of normalized circumferential strain in the thoracic and abdominal aorta using DENSE cardiovascular magnetic resonance. J. Cardiovasc. Magn. Reson. 2019, 21, 59.
- Bracamonte, J.H.; Wilson, J.S.; Soares, J.S. Assessing Patient-Specific Mechanical Properties of Aortic Wall and Peri-Aortic Structures from In Vivo DENSE Magnetic Resonance Imaging Using an Inverse Finite Element Method and Elastic Foundation Boundary Conditions. J. Biomech. Eng. 2020, 142, 1210111–12101113.
- Bonnet, M.; Constantinescu, A. Inverse problems in elasticity. Inverse Probl. 2005, 21, R1–R50.
- Wang, W.; Wang, D.; Falisse, A.; Severijns, P.; Overbergh, T.; Moke, L.; Scheys, L.; De Groote, F.; Jonkers, I. A Dynamic Optimization Approach for Solving Spine Kinematics While Calibrating Subject-Specific Mechanical Properties. Ann. Biomed. Eng. 2021, 49, 2311–2322.
- Zhang, Y.; Wang, V.Y.; Morgan, A.E.; Kim, J.; Ge, L.; Guccione, J.M.; Weinsaft, J.W.; Ratcliffe, M.B. A Novel MRI-Based Finite Element Modeling Method for Calculation of Myocardial Ischemia Effect in Patients with Functional Mitral Regurgitation. Front. Physiol. 2020, 11, 158.
- Avazmohammadi, R.; Li, D.S.; Leahy, T.; Shih, E.; Soares, J.S.; Gorman, J.H.; Gorman, R.C.; Sacks, M.S. An integrated inverse model-experimental approach to determine soft tissue three-dimensional constitutive parameters: Application to post-infarcted myocardium. Biomech. Model. Mechanobiol. 2018, 17, 31–53.
- Fung, Y.C. Elasticity of soft tissues in simple elongation. Am. J. Physiol. Content 1967, 213, 1532–1544.
- Chuong, C.J.; Fung, Y.C. Three-Dimensional Stress Distribution in Arteries. J. Biomech. Eng. 1983, 105, 268–274.
- Pfaller, M.R.; Hörmann, J.M.; Weigl, M.; Nagler, A.; Chabiniok, R.; Bertoglio, C.; Wall, W.A. The importance of the pericardium for cardiac biomechanics: From physiology to computational modeling. Biomech. Model. Mechanobiol. 2019, 18, 503–529.
- Liu, Y.; Dang, C.; Garcia, M.; Gregersen, H.; Kassab, G.S. Surrounding tissues affect the passive mechanics of the vessel wall: Theory and experiment. Am. J. Physiol. Circ. Physiol. 2007, 293, 3290–3300.
- Humphrey, J.D. Mechanics of the Arterial Wall: Review and Directions. Crit. Rev. Biomed. Eng. 1995, 23, 1–162.
- Sokolis, D.P.; Savva, G.D.; Papadodima, S.A.; Kourkoulis, S.K. Regional distribution of circumferential residual strains in the human aorta according to age and gender. J. Mech. Behav. Biomed. Mater. 2017, 67, 87–100.
- Sokolis, D.P.; Bompas, A.; Papadodima, S.A.; Kourkoulis, S.K. Variation of Axial Residual Strains along the Course and Circumference of Human Aorta Considering Age and Gender. J. Biomech. Eng. 2020, 142, 0210031–02100313.
- Cardamone, L.; Valentín, A.; Eberth, J.F.; Humphrey, J.D. Origin of axial prestretch and residual stress in arteries. Biomech. Model. Mechanobiol. 2009, 8, 431–446.
- Holzapfel, G.; Gasser, T.; Stadler, M. A structural model for the viscoelastic behavior of arterial walls: Continuum formulation and finite element analysis. Eur. J. Mech. A Solids 2002, 21, 441–463.
- Humphrey, J.D.; Na, S. Elastodynamics and Arterial Wall Stress. Ann. Biomed. Eng. 2002, 30, 509–523.
- Akyildiz, A.C.; Speelman, L.; Gijsen, F.J.H. Mechanical properties of human atherosclerotic intima tissue. J. Biomech. 2014, 47, 773–783.
- Wang, Z.; Golob, M.J.; Chesler, N.C. Viscoelastic Properties of Cardiovascular Tissues. Viscoelast. Viscoplast. Mater. 2016, 2, 64.
- Barker, A.J.; van Ooij, P.; Bandi, K.; Garcia, J.; Albaghdadi, M.; McCarthy, P.; Bonow, R.O.; Carr, J.; Collins, J.; Malaisrie, S.C.; et al. Viscous energy loss in the presence of abnormal aortic flow. Magn. Reson. Med. 2014, 72, 620–628.
- Mahalingam, A.; Gawandalkar, U.U.; Kini, G.; Buradi, A.; Araki, T.; Ikeda, N.; Nicolaides, A.; Laird, J.R.; Saba, L.; Suri, J.S. Numerical analysis of the effect of turbulence transition on the hemodynamic parameters in human coronary arteries. Cardiovasc. Diagn. Ther. 2016, 6, 208–220.
- Womersley, J.R. Method for the calculation of velocity, rate of flow and viscous drag in arteries when the pressure gradient is known. J. Physiol. 1955, 127, 553–563.
- McDonald, D.A. The relation of pulsatile pressure to flow in arteries. J. Physiol. 1955, 127, 533–552.
- Taylor, M.G. The Discrepancy between Steady- and Oscillatory-Flow Calibration of Flowmeters of the ‘Bristle’and ‘Pendulum’ Types: A Theoretical Study. Phys. Med. Biol. 1958, 2, 324–337.
- Pedley, T.J.; Schroter, R.C.; Sudlow, M.F. Pressure flow relations in branched tubes. J. Physiol. 1969, 204, 114. Available online: https://europepmc.org/article/med/5824621 (accessed on 29 November 2021).
- Williams, M.C.; Rosenblatt, J.S.; Soane, D.S. Theory of Blood Rheology Based on a Statistical Mechanics Treatment of Rouleaux, and Comparisons with Data. Int. J. Polym. Mater. Polym. Biomater. 1993, 21, 57–63.
- Soares, J.S.; Gao, C.; Alemu, Y.; Slepian, M.; Bluestein, D. Simulation of platelets suspension flowing through a stenosis model using a dissipative particle dynamics approach. Ann. Biomed. Eng. 2013, 41, 2318–2333.
- Fedosov, D.A.; Noguchi, H.; Gompper, G. Multiscale modeling of blood flow: From single cells to blood rheology. Biomech. Model. Mechanobiol. 2013, 132, 239–258.
- Peskin, C.S. The immersed boundary method. Acta Numer. 2002, 11, 479–517.
- Flamini, V.; DeAnda, A.; Griffith, B.E. Immersed boundary-finite element model of fluid–structure interaction in the aortic root. Theor. Comput. Fluid Dyn. 2015, 30, 139–164.
- Yang, X.; Zhang, X.; Li, Z.; He, G.W. A smoothing technique for discrete delta functions with application to immersed boundary method in moving boundary simulations. J. Comput. Phys. 2009, 228, 7821–7836.
- van Loon, R.; Anderson, P.D.; van de Vosse, F.; Sherwin, S. Comparison of various fluid–structure interaction methods for deformable bodies. Comput. Struct. 2007, 85, 833–843.
- Moukalled, F.D.F.; Mangani, L. The Finite Volume Method in Computational Fluid Dynamics; Springer: New York, NY, USA, 2015.
- Mackerle, J. Finite element modelling and simulations in cardiovascular mechanics and cardiology: A bibliography 1993–2004. Comput. Methods Biomech. Biomed. Eng. 2005, 8, 59–81.
- Fallah, N.A.; Bailey, C.; Cross, M.; Taylor, G. Comparison of finite element and finite volume methods application in geometrically nonlinear stress analysis. Appl. Math. Model. 2000, 24, 439–455.
- Jeong, W.; Seong, J. Comparison of effects on technical variances of computational fluid dynamics (CFD) software based on finite element and finite volume methods. Int. J. Mech. Sci. 2014, 78, 19–26.
- Sabatier, P.C. Inverse Problems—An introduction. Inverse Probl. 1985, 1, 302.
- Grédiac, M.; Toussaint, E.; Pierron, F. Special virtual fields for the direct determination of material parameters with the virtual fields method. 2—Application to in-plane properties. Int. J. Solids Struct. 2002, 39, 2707–2730.
- Grediac, M.; Toussaint, E.; Pierron, F. Special virtual fields for the direct determination of material parameters with the virtual fields method. 1—Principle and definition. Int. J. Solids Struct. 2002, 39, 2691–2705.
- Tessler, A.; Spangler, J.L. A least-squares variational method for full-field reconstruction of elastic deformations in shear-deformable plates and shells. Comput. Methods Appl. Mech. Eng. 2005, 194, 327–339.
- Beck, J.V.; A Woodbury, K.A. Inverse problems and parameter estimation: Integration of measurements and analysis. Meas. Sci. Technol. 1998, 9, 839–847.
- Lassila, T.; Manzoni, A.; Quarteroni, A.; Rozza, G. A reduced computational and geometrical framework for inverse problems in hemodynamics. Int. J. Numer. Methods Biomed. Eng. 2013, 29, 741–776.
- Liu, M.; Liang, L.; Sun, W. A new inverse method for estimation of in vivo mechanical properties of the aortic wall. J. Mech. Behav. Biomed. Mater. 2017, 72, 148–158.
- Cotter, S.L.; Dashti, M.; Robinson, J.C.; Stuart, A.M. Bayesian inverse problems for functions and applications to fluid mechanics. Inverse Probl. 2009, 25, 115008.
- Wohlfahrt, P.; Krajčoviechová, A.; Seidlerová, J.; Mayer, O.; Bruthans, J.; Filipovský, J.; Laurent, S.; Cífková, R. Arterial stiffness parameters: How do they differ? Atherosclerosis 2013, 231, 359–364.
- Vitarelli, A.; Conde, Y.; Cimino, E.; D’Angeli, I.; D’Orazio, S.; Stellato, S.; Padella, V.; Caranci, F. Aortic Wall Mechanics in the Marfan Syndrome Assessed by Transesophageal Tissue Doppler Echocardiography. Am. J. Cardiol. 2006, 97, 571–577.
- Mack, W.J.; Islam, T.; Lee, Z.; Selzer, R.H.; Hodis, H.N. Environmental tobacco smoke and carotid arterial stiffness. Prev. Med. 2003, 37, 148–154.
- Töger, J.; Zahr, M.J.; Aristokleous, N.; Bloch, K.M.; Carlsson, M.; Persson, P.-O. Blood flow imaging by optimal matching of computational fluid dynamics to 4D-flow data. Magn. Reson. Med. 2020, 84, 2231–2245.
- Hess, A.T.; Bissell, M.M.; Ntusi, N.A.B.; Lewis, A.J.M.; Tunnicliffe, E.M.; Greiser, A.; Stalder, A.F.; Francis, J.M.; Myerson, S.G.; Neubauer, S.; et al. Aortic 4D flow: Quantification of signal-to-noise ratio as a function of field strength and contrast enhancement for 1.5 T, 3 T, and 7 T. Magn. Reson. Med. 2015, 73, 1864–1871.
- Aletras, A.H.; Ding, S.; Balaban, R.S.; Wen, H. DENSE: Displacement Encoding with Stimulated Echoes in Cardiac Functional MRI. J. Magn. Reson. 1999, 137, 247–252.
- Bracamonte, J.H.; Wilson, J.S.; Soares, J.S. Quantification of the heterogeneous effect of static and dynamic perivascular struc-tures on patient-specific local aortic wall mechanics using inverse finite element modeling and DENSE MRI. J. Biomech. 2009, 11, 123.
- Busch, J.; Giese, D.; Kozerke, S. Image-based background phase error correction in 4D flow MRI revisited. J. Magn. Reson. Imaging 2017, 46, 1516–1525.
- Ng, A.; Swanevelder, J. Resolution in ultrasound imaging. Contin. Educ. Anaesth. Crit. Care Pain 2011, 11, 186–192.
- Sassaroli, E.; Crake, C.; Scorza, A.; Kim, D.; Park, M. Image quality evaluation of ultrasound imaging systems: Advanced B-modes. J. Appl. Clin. Med. Phys. 2019, 20, 115–124.
- Linte, C.A.; Moore, J.; Wiles, A.D.; Wedlake, C.; Peters, T.M. Virtual reality-enhanced ultrasound guidance: A novel technique for intracardiac interventions. Comput. Aided Surg. 2008, 13, 82–94.
- Olson, I.; Brabender, J.; Thorson, K.; Lopez, L. 3D Echocardiography. In Multimodality Imaging Innovations in Adult Congenital Heart Disease; Springer Nature: Berlin, Germany, 2021; pp. 3–25.
- Malik, S.B.; Chen, N.; Parker, R.A.; Hsu, J.Y. Transthoracic Echocardiography: Pitfalls and Limitations as Delineated at Cardiac CT and MR Imaging. RadioGraphics 2017, 37, 383–406.
- Ohayon, J.; Finet, G.; Le Floc’H, S.; Cloutier, G.; Gharib, A.; Heroux, J.; Pettigrew, R.I. Biomechanics of Atherosclerotic Coronary Plaque: Site, Stability and In Vivo Elasticity Modeling. Ann. Biomed. Eng. 2014, 42, 269–279.
- Ko, S.M.; Hwang, S.H.; Lee, H.-J. Role of Cardiac Computed Tomography in the Diagnosis of Left Ventricular Myocardial Diseases. J. Cardiovasc. Imaging 2019, 27, 73–92.
- Truong, U.T.; Kutty, S.; Broberg, C.S.; Sahn, D.J. Multimodality Imaging in Congenital Heart Disease: An Update. Curr. Cardiovasc. Imaging Rep. 2012, 5, 481–490.
- Campbell-Washburn, A.E.; Tavallaei, M.A.; Pop, M.; Grant, E.K.; Chubb, H.; Rhode, K.; Wright, G.A. Real-time MRI guidance of cardiac interventions. J. Magn. Reson. Imaging 2017, 46, 935–950.
- Pop, M.; Ghugre, N.R.; Ramanan, V.; Morikawa, L.; Stanisz, G.; Dick, A.J.; Wright, G.A. Quantification of fibrosis in infarcted swine hearts byex vivolate gadolinium-enhancement and diffusion-weighted MRI methods. Phys. Med. Biol. 2013, 58, 5009–5028.
- Dall’Armellina, E.; Bissell, M.M.; Broadbent, D.A.; Plein, S. MRI T1 Mapping: Myocardial Fibrosis. In Multimodality Imaging Innovations in Adult Congenital Heart Disease; Springer Nature: Berlin, Germany, 2021; pp. 49–61.
- Kwon, S.T.; Burek, W.; Dupay, A.C.; Farsad, M.; Baek, S.; Park, E.-A.; Lee, W. Interaction of expanding abdominal aortic aneurysm with surrounding tissue: Retrospective CT image studies. S. Pac. J. Nat. Appl. Sci. 2015, 1, e150.
- Kofler, J.M.; Cody, D.D.; Morin, R.L. CT Protocol Review and Optimization. J. Am. Coll. Radiol. 2014, 11, 267–270.
- Maas, S.A.; Erdemir, A.; Halloran, J.P.; Weiss, J.A. A general framework for application of prestrain to computational models of biological materials. J. Mech. Behav. Biomed. Mater. 2016, 61, 499–510.
- Nikou, A.; Dorsey, S.M.; McGarvey, J.R.; Gorman, J.H.; Burdick, J.A.; Pilla, J.J.; Gorman, R.C.; Wenk, J.F. Effects of using the unloaded configuration in predicting the in vivo diastolic properties of the heart. Comput. Methods Biomech. Biomed. Eng. 2016, 19, 1714–1720.
- Bols, J.; Degroote, J.; Trachet, B.; Verhegghe, B.; Segers, P.; Vierendeels, J. A computational method to assess the in vivo stresses and unloaded configuration of patient-specific blood vessels. J. Comput. Appl. Math. 2013, 246, 10–17.
- Xi, J.; Lamata, P.; Niederer, S.; Land, S.; Shi, W.; Zhuang, X.; Ourselin, S.; Duckett, S.G.; Shetty, A.K.; Rinaldi, C.A.; et al. The estimation of patient-specific cardiac diastolic functions from clinical measurements. Med. Image Anal. 2013, 17, 133–146.
- Finsberg, H.; Balaban, G.; Ross, S.; Håland, T.F.; Odland, H.H.; Sundnes, J.; Wall, S. Estimating cardiac contraction through high resolution data assimilation of a personalized mechanical model. J. Comput. Sci. 2018, 24, 85–90.
- Aguado-Sierra, J.; Krishnamurthy, A.; Villongco, C.; Chuang, J.; Howard, E.; Gonzales, M.J.; Omens, J.; Krummen, D.E.; Narayan, S.; Kerckhoffs, R.C.P.; et al. Patient-specific modeling of dyssynchronous heart failure: A case study. Prog. Biophys. Mol. Biol. 2011, 107, 147–155.
- Sermesant, M.; Moireau, P.; Camara, O.; Sainte-Marie, J.; Andriantsimiavona, R.; Cimrman, R.; Hill, D.L.G.; Chapelle, D.; Razavi, R. Cardiac function estimation from MRI using a heart model and data assimilation: Advances and difficulties. Med. Image Anal. 2006, 10, 642–656.
- Balaban, G.; Finsberg, H.; Funke, S.; Håland, T.F.; Hopp, E.; Sundnes, J.; Wall, S.; Rognes, M.E. In vivo estimation of elastic heterogeneity in an infarcted human heart. Biomech. Model. Mechanobiol. 2018, 17, 1317–1329.
- Rumindo, G.K.; Ohayon, J.; Croisille, P.; Clarysse, P. In vivo estimation of normal left ventricular stiffness and contractility based on routine cine MR acquisition. Med. Eng. Phys. 2020, 85, 16–26.
- Wang, Z.J.; Wang, V.Y.; Bradley, C.P.; Nash, M.P.; Young, A.A.; Cao, J.J. Left Ventricular Diastolic Myocardial Stiffness and End-Diastolic Myofibre Stress in Human Heart Failure Using Personalised Biomechanical Analysis. J. Cardiovasc. Transl. Res. 2018, 11, 346–356.
- Finsberg, H.; Xi, C.; Zhao, X.; Le Tan, J.; Genet, M.; Sundnes, J.S.; Lee, L.C.; Zhong, L.; Wall, S.T. Computational quantification of patient-specific changes in ventricular dynamics associated with pulmonary hypertension. Am. J. Physiol. Circ. Physiol. 2019, 317, H1363–H1375.
- Asner, L.; Hadjicharalambous, M.; Chabiniok, R.; Peressutti, D.; Sammut, E.; Wong, J.; Carr-White, G.; Razavi, R.; King, A.P.; Smith, N.; et al. Patient-specific modeling for left ventricular mechanics using data-driven boundary energies. Comput. Methods Appl. Mech. Eng. 2016, 314, 269–295.
- Nasopoulou, A.; Shetty, A.; Lee, J.; Nordsletten, D.; Rinaldi, C.; Lamata, P.; Niederer, S. Improved identifiability of myocardial material parameters by an energy-based cost function. Biomech. Model. Mechanobiol. 2017, 16, 971–988.
- Gao, H.; Aderhold, A.; Mangion, K.; Luo, X.; Husmeier, D.; Berry, C. Changes and classification in myocardial contractile function in the left ventricle following acute myocardial infarction. J. R. Soc. Interface 2017, 14, 20170203.
- Marchesseau, S.; Delingette, H.; Sermesant, M.; Cabrera-Lozoya, R.; Tobon-Gomez, C.; Moireau, P.; Ventura, R.F.; Lekadir, K.; Hernandez, A.; Garreau, M.; et al. Personalization of a cardiac electromechanical model using reduced order unscented Kalman filtering from regional volumes. Med. Image Anal. 2013, 17, 816–829.
- Xi, J.; Lamata, P.; Shi, W.; Niederer, S.; Land, S.; Rueckert, D.; Duckett, S.G.; Shetty, A.K.; Rinaldi, C.A.; Razavi, R.; et al. An Automatic Data Assimilation Framework for Patient-Specific Myocardial Mechanical Parameter Estimation. In Functional Imaging and Modeling of the Heart; Springer: Berlin/Heidelberg, Germany, 2011; pp. 392–400.
- Misfeld, M.; Sievers, H.-H. Heart valve macro- and microstructure. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 1421–1436.
- Lee, C.-H.; Laurence, D.W.; Ross, C.J.; Kramer, K.E.; Babu, A.R.; Johnson, E.L.; Hsu, M.-C.; Aggarwal, A.; Mir, A.; Burkhart, H.M.; et al. Mechanics of the Tricuspid Valve—From Clinical Diagnosis/Treatment, In-Vivo and In-Vitro Investigations, to Patient-Specific Biomechanical Modeling. Bioengineering 2019, 6, 47.
- Krishnamurthy, G.; Ennis, D.B.; Itoh, A.; Bothe, W.; Swanson, J.C.; Karlsson, M.; Kuhl, E.; Miller, D.C.; Ingels, N.B. Material properties of the ovine mitral valve anterior leaflet in vivo from inverse finite element analysis. Am. J. Physiol. Circ. Physiol. 2008, 295, H1141–H1149.
- Itoh, A.; Krishnamurthy, G.; Swanson, J.C.; Ennis, D.B.; Bothe, W.; Kuhl, E.; Karlsson, M.; Davis, L.R.; Miller, D.C.; Ingels, N.B. Active stiffening of mitral valve leaflets in the beating heart. Am. J. Physiol. Circ. Physiol. 2009, 296, H1766–H1773.
- Lee, C.-H.; Amini, R.; Gorman, R.; Gorman, J.; Sacks, M. An inverse modeling approach for stress estimation in mitral valve anterior leaflet valvuloplasty for in-vivo valvular biomaterial assessment. J. Biomech. 2014, 47, 2055–2063.
- Lee, C.-H.; Zhang, W.; Feaver, K.; Gorman, R.C.; Gorman, J.H.; Sacks, M.S. On the in vivo function of the mitral heart valve leaflet: Insights into tissue–interstitial cell biomechanical coupling. Biomech. Model. Mechanobiol. 2017, 16, 1613–1632.
- Aggarwal, A.; Pouch, A.M.; Lai, E.; Lesicko, J.; Yushkevich, P.A.; Gorman, J.H.; Gorman, R.C.; Sacks, M.S. In-vivo heterogeneous functional and residual strains in human aortic valve leaflets. J. Biomech. 2016, 49, 2481–2490.
- Franquet, A.; Avril, S.; Le Riche, R.; Badel, P.; Schneider, F.C.; Li, Z.Y.; Boissier, C.; Favre, J.P. A New Method for the In Vivo Identification of Mechanical Properties in Arteries from Cine MRI Images: Theoretical Framework and Validation. IEEE Trans. Med. Imaging 2013, 32, 1448–1461.
- Wittek, A.; Karatolios, K.; Bihari, P.; Schmitz-Rixen, T.; Moosdorf, R.; Vogt, S.; Blase, C. In vivo determination of elastic properties of the human aorta based on 4D ultrasound data. J. Mech. Behav. Biomed. Mater. 2013, 27, 167–183.
- Wittek, A.; Derwich, W.; Karatolios, K.; Fritzen, C.P.; Vogt, S.; Schmitz-Rixen, T.; Blase, C. A finite element updating approach for identification of the anisotropic hyperelastic properties of normal and diseased aortic walls from 4D ultrasound strain imaging. J. Mech. Behav. Biomed. Mater. 2016, 58, 122–138.
- Karatolios, K.; Wittek, A.; Nwe, T.H.; Bihari, P.; Shelke, A.; Josef, D.; Schmitz-Rixen, T.; Geks, J.; Maisch, B.; Blase, C.; et al. Method for Aortic Wall Strain Measurement with Three-Dimensional Ultrasound Speckle Tracking and Fitted Finite Element Analysis. Ann. Thorac. Surg. 2013, 96, 1664–1671.
- Xi, J.; Lamata, P.; Lee, J.; Moireau, P.; Chapelle, D.; Smith, N. Myocardial transversely isotropic material parameter estimation from in-silico measurements based on a reduced-order unscented Kalman filter. J. Mech. Behav. Biomed. Mater. 2011, 4, 1090–1102.
- Van Disseldorp, E.M.J.; Petterson, N.J.; Van De Vosse, F.N.; Van Sambeek, M.R.H.M.; Lopata, R.G.P. Quantification of aortic stiffness and wall stress in healthy volunteers and abdominal aortic aneurysm patients using time-resolved 3D ultrasound: A comparison study. Eur. Heart J.—Cardiovasc. Imaging 2019, 20, 185–191.
- Talou, G.D.M.; Blanco, P.J.; Ares, G.D.; Bezerra, C.G.; Lemos, P.A.; Feijóo, R.A. Mechanical Characterization of the Vessel Wall by Data Assimilation of Intravascular Ultrasound Studies. Front. Physiol. 2018, 9, 292.
- Wang, Q.; Canton, G.; Guo, J.; Guo, X.; Hatsukami, T.S.; Billiar, K.L.; Yuan, C.; Wu, Z.; Tang, D. MRI-based patient-specific human carotid atherosclerotic vessel material property variations in patients, vessel location and long-term follow up. PLoS ONE 2017, 12, e0180829.
- Liu, H.; Canton, G.; Yuan, C.; Yang, C.; Billiar, K.; Teng, Z.; Hoffman, A.H.; Tang, D. Using In Vivo Cine and 3D Multi-Contrast MRI to Determine Human Atherosclerotic Carotid Artery Material Properties and Circumferential Shrinkage Rate and Their Impact on Stress/Strain Predictions. J. Biomech. Eng. 2012, 134, 011008–0110089.
- Bracamonte, J.; Wilson, J.S.; Soares, J.S. Modeling Patient-Specific Periaortic Interactions with Static and Dynamic Structures Using a Moving Heterogeneous Elastic Foundation Boundary Condition. In Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2021; pp. 315–327.
- Quick, C.M.; Berger, D.S.; Stewart, R.H.; Laine, G.A.; Hartley, C.; Noordergraaf, A. Resolving the Hemodynamic Inverse Problem. IEEE Trans. Biomed. Eng. 2006, 53, 361–368.
- Pant, S.; Corsini, C.; Baker, C.; Hsia, T.-Y.; Pennati, G.; Vignon-Clementel, I.E. Inverse problems in reduced order models of cardiovascular haemodynamics: Aspects of data assimilation and heart rate variability. J. R. Soc. Interface 2017, 14, 20160513.
- Quick, C.M.; Young, W.L.; Noordergraaf, A. Infinite number of solutions to the hemodynamic inverse problem. Am. J. Physiol. Circ. Physiol. 2001, 280, H1472–H1479.
- Rispoli, V.C.; Nielsen, J.F.; Nayak, K.S.; Carvalho, J.L.A. Computational fluid dynamics simulations of blood flow regularized by 3D phase contrast MRI. Biomed. Eng. Online 2015, 14, 1–23.
- Moireau, P.; Chapelle, D. Reduced-order Unscented Kalman Filtering with application to parameter identification in large-dimensional systems. ESAIM Control. Optim. Calc. Var. 2011, 17, 380–405.





