
Video Upload Options
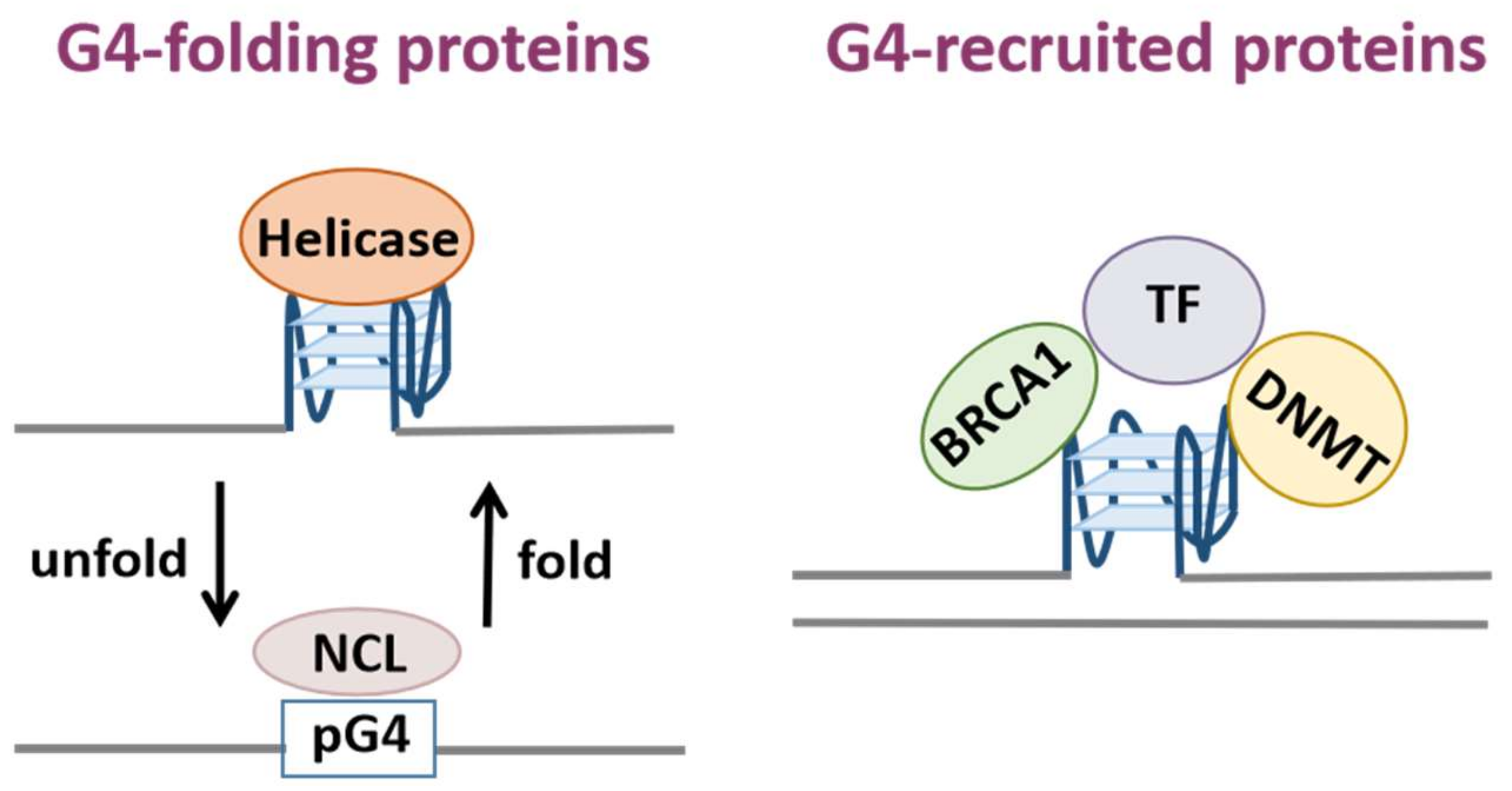
G-quadruplexes (G4s) are non-canonical secondary nucleic acid structures. Sequences with the potential to form G4s are abundant in regulatory regions of the genome including telomeres, promoters and 5′ non-coding regions, indicating they fulfill important genome regulatory functions. In recent years, an increasing number of G-quadruplex-binding proteins have been identified with biochemical experiments. G4-binding proteins are involved in vital cellular processes such as telomere maintenance, DNA replication, gene transcription, mRNA processing. Therefore, G4-binding proteins are also associated with various human diseases.
1. G-Quadruplex-Binding Proteins
1.1. Detection of G-Quadruplex-Binding Proteins
1.2. DNA G-Quadruplex-Binding Proteins
1.2.1. Telomeric G-Quadruplex-Binding Proteins
1.2.2. G-Quadruplex-Binding Proteins Involved in Replication
1.2.3. G-Quadruplex-Binding Proteins Involved in Transcription
1.2.4. Other DNA G-Quadruplex-Binding Proteins
1.3. RNA G-Quadruplex-Binding Proteins
2. Structural Properties of G-Quadruplex-Binding Proteins
2.1. RGG Domain
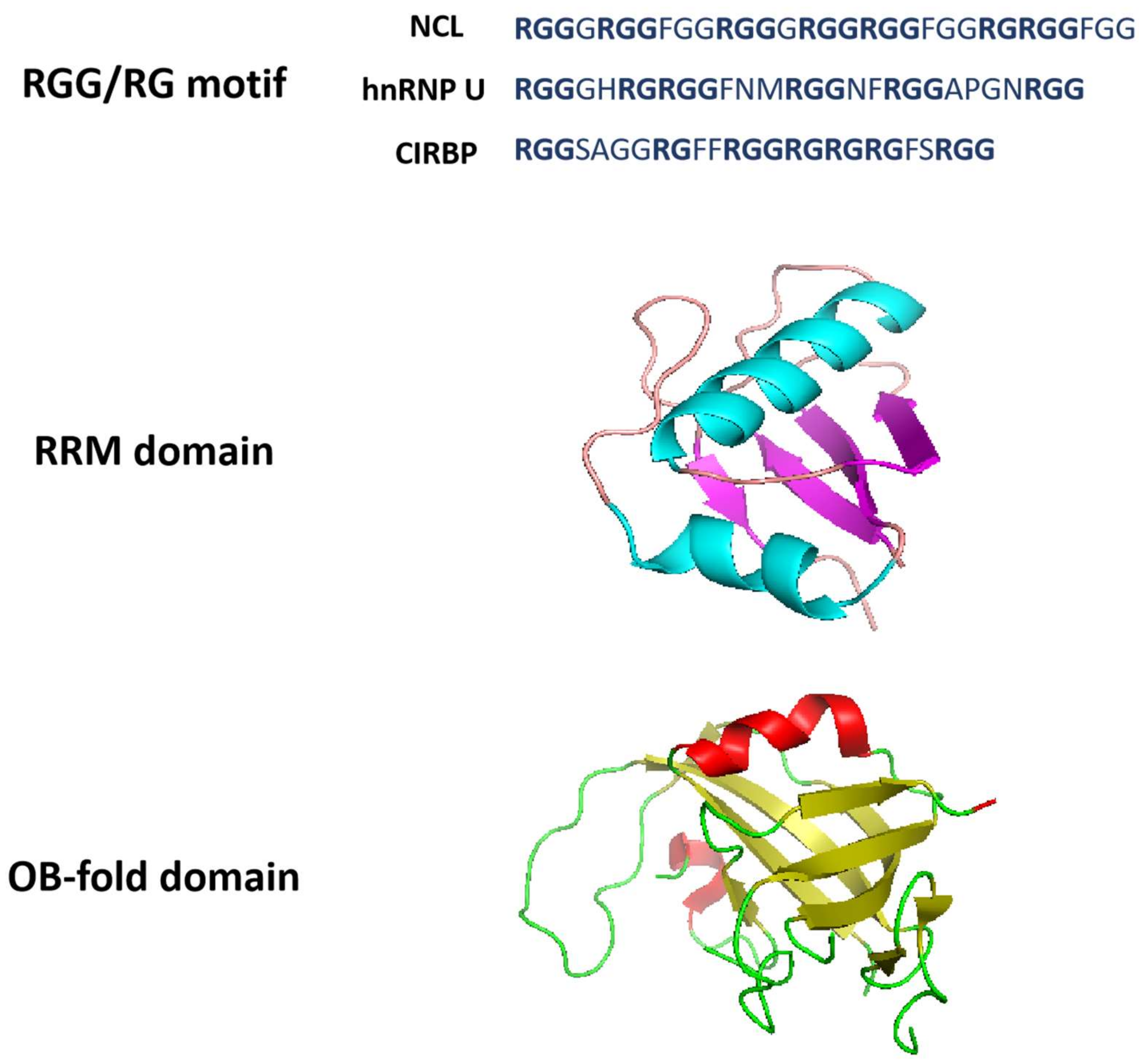
Figure 2. Structural properties of G-quadruplex-binding proteins. RGG/RG motifs are from NCL, hnRNP U and CIRBP. The RRM domain structure is derived from Protein Data Bank with structure code 2KRR (NCL). RRM domain is an αβ sandwich structure composed of one four-stranded antiparallel β-sheet and two α-helices packed against the β-sheet. The OB-fold domain structure is derived from Protein Data Bank with structure code 5W2L (CTC1). OB-fold domain is a β-barrel formed by five antiparallel β-sheets.
2.2. RRM Domain
2.3. OB-Fold Domain
References
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136.
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474.
- Matsumoto, K.; Okamoto, K.; Okabe, S.; Fujii, R.; Ueda, K.; Ohashi, K.; Seimiya, H. G-quadruplex-forming nucleic acids interact with splicing factor 3B subunit 2 and suppress innate immune gene expression. Genes Cells 2021, 26, 65–82.
- Serikawa, T.; Spanos, C.; von Hacht, A.; Budisa, N.; Rappsilber, J.; Kurreck, J. Comprehensive identification of proteins binding to RNA G-quadruplex motifs in the 5′ UTR of tumor-associated mRNAs. Biochimie 2018, 144, 169–184.
- Lee, C.Y.; McNerney, C.; Myong, S. G-Quadruplex and Protein Binding by Single-Molecule FRET Microscopy. Methods Mol. Biol. 2019, 2035, 309–322.
- Scalabrin, M.; Frasson, I.; Ruggiero, E.; Perrone, R.; Tosoni, E.; Lago, S.; Tassinari, M.; Palu, G.; Richter, S.N. The cellular protein hnRNP A2/B1 enhances HIV-1 transcription by unfolding LTR promoter G-quadruplexes. Sci. Rep. 2017, 7, 45244.
- McRae, E.K.S.; Booy, E.P.; Padilla-Meier, G.P.; McKenna, S.A. On Characterizing the Interactions between Proteins and Guanine Quadruplex Structures of Nucleic Acids. J. Nucleic Acids 2017, 2017, 9675348.
- Brazda, V.; Cerven, J.; Bartas, M.; Mikyskova, N.; Coufal, J.; Pecinka, P. The Amino Acid Composition of Quadruplex Binding Proteins Reveals a Shared Motif and Predicts New Potential Quadruplex Interactors. Molecules 2018, 23, 2341.
- Huang, Z.L.; Dai, J.; Luo, W.H.; Wang, X.G.; Tan, J.H.; Chen, S.B.; Huang, Z.S. Identification of G-Quadruplex-Binding Protein from the Exploration of RGG Motif/G-Quadruplex Interactions. J. Am. Chem Soc. 2018, 140, 17945–17955.
- Zhang, X.; Spiegel, J.; Martinez Cuesta, S.; Adhikari, S.; Balasubramanian, S. Chemical profiling of DNA G-quadruplex-interacting proteins in live cells. Nat. Chem. 2021, 13, 626–633.
- Brazda, V.; Haronikova, L.; Liao, J.C.; Fojta, M. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014, 15, 17493–17517.
- Izumi, H.; Funa, K. Telomere Function and the G-Quadruplex Formation are Regulated by hnRNP U. Cells 2019, 8, 390.
- Takahama, K.; Kino, K.; Arai, S.; Kurokawa, R.; Oyoshi, T. Identification of Ewing’s sarcoma protein as a G-quadruplex DNA- and RNA-binding protein. FEBS J. 2011, 278, 988–998.
- Takahama, K.; Takada, A.; Tada, S.; Shimizu, M.; Sayama, K.; Kurokawa, R.; Oyoshi, T. Regulation of Telomere Length by G-Quadruplex Telomere DNA- and TERRA-Binding Protein TLS/FUS. Chem. Biol. 2013, 20, 341–350.
- Chen, Y. The structural biology of the shelterin complex. Biol. Chem. 2019, 400, 457–466.
- Stewart, J.A.; Chaiken, M.F.; Wang, F.; Price, C.M. Maintaining the end: Roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat. Res.-Fund. Mol. M 2012, 730, 12–19.
- Arnoult, N.; Karlseder, J. Complex interactions between the DNA-damage response and mammalian telomeres. Nat. Struct. Mol. Biol. 2015, 22, 859–866.
- Baumann, P.; Cech, T.R. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 2001, 292, 1171–1175.
- Wang, F.; Podell, E.R.; Zaug, A.J.; Yang, Y.; Baciu, P.; Cech, T.R.; Lei, M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 2007, 445, 506–510.
- Chaires, J.B.; Gray, R.D.; Dean, W.L.; Monsen, R.; DeLeeuw, L.W.; Stribinskis, V.; Trent, J.O. Human POT1 unfolds G-quadruplexes by conformational selection. Nucleic Acids Res. 2020, 48, 4976–4991.
- Bhattacharjee, A.; Wang, Y.; Diao, J.; Price, C.M. Dynamic DNA binding, junction recognition and G4 melting activity underlie the telomeric and genome-wide roles of human CST. Nucleic Acids Res. 2017, 45, 12311–12324.
- Wu, W.; Rokutanda, N.; Takeuchi, J.; Lai, Y.; Maruyama, R.; Togashi, Y.; Nishikawa, H.; Arai, N.; Miyoshi, Y.; Suzuki, N.; et al. HERC2 Facilitates BLM and WRN Helicase Complex Interaction with RPA to Suppress G-Quadruplex DNA. Cancer Res. 2018, 78, 6371–6385.
- Budhathoki, J.B.; Ray, S.; Urban, V.; Janscak, P.; Yodh, J.G.; Balci, H. RecQ-core of BLM unfolds telomeric G-quadruplex in the absence of ATP. Nucleic Acids Res. 2014, 42, 11528–11545.
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637.
- Miyake, Y.; Nakamura, M.; Nabetani, A.; Shimamura, S.; Tamura, M.; Yonehara, S.; Saito, M.; Ishikawa, F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 2009, 36, 193–206.
- Zhang, M.M.; Wang, B.; Li, T.F.; Liu, R.; Xiao, Y.N.; Geng, X.; Li, G.; Liu, Q.; Price, C.M.; Liu, Y.; et al. Mammalian CST averts replication failure by preventing G-quadruplex accumulation. Nucleic Acids Res. 2019, 47, 5243–5259.
- Surovtseva, Y.V.; Churikov, D.; Boltz, K.A.; Song, X.Y.; Lamb, J.C.; Warrington, R.; Leehy, K.; Heacock, M.; Price, C.M.; Shippen, D.E. Conserved Telomere Maintenance Component 1 Interacts with STN1 and Maintains Chromosome Ends in Higher Eukaryotes. Mol. Cell 2009, 36, 207–218.
- Hwang, H.; Buncher, N.; Opresko, P.L.; Myong, S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure 2012, 20, 1872–1880.
- Leon-Ortiz, A.M.; Svendsen, J.; Boulton, S.J. Metabolism of DNA secondary structures at the eukaryotic replication fork. DNA Repair 2014, 19, 152–162.
- Valton, A.L.; Prioleau, M.N. G-Quadruplexes in DNA Replication: A Problem or a Necessity? Trends Genet. 2016, 32, 697–706.
- Sauer, M.; Paeschke, K. G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 2017, 45, 1173–1182.
- Sun, Z.Y.; Wang, X.N.; Cheng, S.Q.; Su, X.X.; Ou, T.M. Developing Novel G-Quadruplex Ligands: From Interaction with Nucleic Acids to Interfering with Nucleic Acid(-)Protein Interaction. Molecules 2019, 24, 396.
- Castillo Bosch, P.; Segura-Bayona, S.; Koole, W.; van Heteren, J.T.; Dewar, J.M.; Tijsterman, M.; Knipscheer, P. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014, 33, 2521–2533.
- Sarkies, P.; Murat, P.; Phillips, L.G.; Patel, K.J.; Balasubramanian, S.; Sale, J.E. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012, 40, 1485–1498.
- Eddy, S.; Ketkar, A.; Zafar, M.K.; Maddukuri, L.; Choi, J.Y.; Eoff, R.L. Human Rev1 polymerase disrupts G-quadruplex DNA. Nucleic Acids Res. 2014, 42, 3272–3285.
- Mendoza, O.; Bourdoncle, A.; Boule, J.B.; Brosh, R.M., Jr.; Mergny, J.L. G-quadruplexes and helicases. Nucleic Acids Res. 2016, 44, 1989–2006.
- Suhasini, A.N.; Brosh, R.M. Fanconi anemia and Bloom’s syndrome crosstalk through FANCJ-BLM helicase interaction. Trends Genet. 2012, 28, 7–13.
- Schwindt, E.; Paeschke, K. Mms1 is an assistant for regulating G-quadruplex DNA structures. Curr. Genet. 2018, 64, 535–540.
- Kim, N. The Interplay between G-quadruplex and Transcription. Curr. Med. Chem. 2019, 26, 2898–2917.
- Spiegel, J.; Cuesta, S.M.; Adhikari, S.; Hansel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 2021, 22, 117.
- Lago, S.; Nadai, M.; Cernilogar, F.M.; Kazerani, M.; Dominiguez Moreno, H.; Schotta, G.; Richter, S.N. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun. 2021, 12, 3885.
- Mishra, S.K.; Tawani, A.; Mishra, A.; Kumar, A. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci. Rep. 2016, 6, 38144.
- Cogoi, S.; Paramasivam, M.; Membrino, A.; Yokoyama, K.K.; Xodo, L.E. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010, 285, 22003–22016.
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413.
- Eddy, J.; Maizels, N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006, 34, 3887–3896.
- Simonsson, T.; Pecinka, P.; Kubista, M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998, 26, 1167–1172.
- Gonzalez, V.; Guo, K.; Hurley, L.; Sun, D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009, 284, 23622–23635.
- Sun, D.; Guo, K.; Rusche, J.J.; Hurley, L.H. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005, 33, 6070–6080.
- Cogoi, S.; Xodo, L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006, 34, 2536–2549.
- Dexheimer, T.S.; Sun, D.; Hurley, L.H. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006, 128, 5404–5415.
- Rankin, S.; Reszka, A.P.; Huppert, J.; Zloh, M.; Parkinson, G.N.; Todd, A.K.; Ladame, S.; Balasubramanian, S.; Neidle, S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005, 127, 10584–10589.
- Qin, Y.; Fortin, J.S.; Tye, D.; Gleason-Guzman, M.; Brooks, T.A.; Hurley, L.H. Molecular cloning of the human platelet-derived growth factor receptor beta (PDGFR-beta) promoter and drug targeting of the G-quadruplex-forming region to repress PDGFR-beta expression. Biochemistry 2010, 49, 4208–4219.
- Palumbo, S.L.; Ebbinghaus, S.W.; Hurley, L.H. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J. Am. Chem Soc. 2009, 131, 10878–10891.
- Thakur, R.K.; Kumar, P.; Halder, K.; Verma, A.; Kar, A.; Parent, J.L.; Basundra, R.; Kumar, A.; Chowdhury, S. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009, 37, 172–183.
- Petr, M.; Helma, R.; Polaskova, A.; Krejci, A.; Dvorakova, Z.; Kejnovska, I.; Navratilova, L.; Adamik, M.; Vorlickova, M.; Brazdova, M. Wild-type p53 binds to MYC promoter G-quadruplex. Biosci. Rep. 2016, 36, e00397.
- Quante, T.; Otto, B.; Brazdova, M.; Kejnovska, I.; Deppert, W.; Tolstonog, G.V. Mutant p53 is a transcriptional co-factor that binds to G-rich regulatory regions of active genes and generates transcriptional plasticity. Cell Cycle 2012, 11, 3290–3303.
- Hansel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272.
- Guilbaud, G.; Murat, P.; Recolin, B.; Campbell, B.C.; Maiter, A.; Sale, J.E.; Balasubramanian, S. Local epigenetic reprogramming induced by G-quadruplex ligands. Nat. Chem. 2017, 9, 1110–1117.
- Zyner, K.G.; Simeone, A.; Flynn, S.M.; Doyle, C.; Marsico, G.; Adhikari, S.; Portella, G.; Tannahill, D.; Balasubramanian, S. G-quadruplex DNA structures in human stem cells and differentiation. Nat. Commun. 2022, 13, 142.
- Hou, Y.; Li, F.Y.; Zhang, R.X.; Li, S.; Liu, H.D.; Qin, Z.H.S.; Sun, X. Integrative characterization of G-Quadruplexes in the three-dimensional chromatin structure. Epigenetics-Us 2019, 14, 894–911.
- Law, M.J.; Lower, K.M.; Voon, H.P.; Hughes, J.R.; Garrick, D.; Viprakasit, V.; Mitson, M.; De Gobbi, M.; Marra, M.; Morris, A.; et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 2010, 143, 367–378.
- Wang, Y.; Yang, J.; Wild, A.T.; Wu, W.H.; Shah, R.; Danussi, C.; Riggins, G.J.; Kannan, K.; Sulman, E.P.; Chan, T.A.; et al. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat. Commun. 2019, 10, 943.
- Cayrou, C.; Coulombe, P.; Vigneron, A.; Stanojcic, S.; Ganier, O.; Peiffer, I.; Rivals, E.; Puy, A.; Laurent-Chabalier, S.; Desprat, R.; et al. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 2011, 21, 1438–1449.
- Besnard, E.; Babied, A.; Lapasset, L.; Milhavet, O.; Parrinello, H.; Dantec, C.; Marin, J.M.; Lemaitre, J.M. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012, 19, 837–844.
- Mao, S.Q.; Ghanbarian, A.T.; Spiegel, J.; Martinez Cuesta, S.; Beraldi, D.; Di Antonio, M.; Marsico, G.; Hansel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. DNA G-quadruplex structures mold the DNA methylome. Nat. Struct. Mol. Biol. 2018, 25, 951–957.
- Tikhonova, P.; Pavlova, I.; Isaakova, E.; Tsvetkov, V.; Bogomazova, A.; Vedekhina, T.; Luzhin, A.V.; Sultanov, R.; Severov, V.; Klimina, K.; et al. DNA G-Quadruplexes Contribute to CTCF Recruitment. Int. J. Mol. Sci. 2021, 22, 7090.
- Herdy, B.; Mayer, C.; Varshney, D.; Marsico, G.; Murat, P.; Taylor, C.; D’Santos, C.; Tannahill, D.; Balasubramanian, S. Analysis of NRAS RNA G-quadruplex binding proteins reveals DDX3X as a novel interactor of cellular G-quadruplex containing transcripts. Nucleic Acids Res. 2018, 46, 11592–11604.
- Kwok, C.K.; Marsico, G.; Sahakyan, A.B.; Chambers, V.S.; Balasubramanian, S. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods 2016, 13, 841–844.
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014, 6, 75–80.
- Agarwala, P.; Pandey, S.; Maiti, S. The tale of RNA G-quadruplex. Org. Biomol. Chem. 2015, 13, 5570–5585.
- Schaeffer, C.; Bardoni, B.; Mandel, J.L.; Ehresmann, B.; Ehresmann, C.; Moine, H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001, 20, 4803–4813.
- Bensaid, M.; Melko, M.; Bechara, E.G.; Davidovic, L.; Berretta, A.; Catania, M.V.; Gecz, J.; Lalli, E.; Bardoni, B. FRAXE-associated mental retardation protein (FMR2) is an RNA-binding protein with high affinity for G-quartet RNA forming structure. Nucleic Acids Res. 2009, 37, 1269–1279.
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An Architectural Role for a Nuclear Noncoding RNA: NEAT1 RNA Is Essential for the Structure of Paraspeckles. Mol. Cell 2009, 33, 717–726.
- Simko, E.A.J.; Liu, H.; Zhang, T.; Velasquez, A.; Teli, S.; Haeusler, A.R.; Wang, J. G-quadruplexes offer a conserved structural motif for NONO recruitment to NEAT1 architectural lncRNA. Nucleic Acids Res. 2020, 48, 7421–7438.
- Tassinari, M.; Richter, S.N.; Gandellini, P. Biological relevance and therapeutic potential of G-quadruplex structures in the human noncoding transcriptome. Nucleic Acids Res. 2021, 49, 3617–3633.
- Thandapani, P.; O’Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG motif. Mol. Cell 2013, 50, 613–623.
- Kharel, P.; Becker, G.; Tsvetkov, V.; Ivanov, P. Properties and biological impact of RNA G-quadruplexes: From order to turmoil and back. Nucleic Acids Res. 2020, 48, 12534–12555.
- Masuzawa, T.; Oyoshi, T. Roles of the RGG Domain and RNA Recognition Motif of Nucleolin in G-Quadruplex Stabilization. ACS Omega 2020, 5, 5202–5208.
- Arumugam, S.; Miller, M.C.; Maliekal, J.; Bates, P.J.; Trent, J.O.; Lane, A.N. Solution structure of the RBD1,2 domains from human nucleolin. J. Biomol. Nmr 2010, 47, 79–83.
- Clery, A.; Blatter, M.; Allain, F.H. RNA recognition motifs: Boring? Not quite. Curr. Opin. Struct. Biol 2008, 18, 290–298.
- Maris, C.; Dominguez, C.; Allain, F.H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131.
- Oyoshi, T.; Masuzawa, T. Modulation of histone modifications and G-quadruplex structures by G-quadruplex-binding proteins. Biochem. Biophys. Res. Commun. 2020, 531, 39–44.
- Ding, J.; Hayashi, M.K.; Zhang, Y.; Manche, L.; Krainer, A.R.; Xu, R.M. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999, 13, 1102–1115.
- Enokizono, Y.; Konishi, Y.; Nagata, K.; Ouhashi, K.; Uesugi, S.; Ishikawa, F.; Katahira, M. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J. Biol. Chem. 2005, 280, 18862–18870.
- Williams, P.; Li, L.; Dong, X.; Wang, Y. Identification of SLIRP as a G Quadruplex-Binding Protein. J. Am. Chem. Soc. 2017, 139, 12426–12429.
- Murzin, A.G. OB(oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 1993, 12, 861–867.
- Nguyen, D.D.; Kim, E.Y.; Sang, P.B.; Chai, W. Roles of OB-Fold Proteins in Replication Stress. Front. Cell Dev. Biol. 2020, 8, 574466.
- Lim, C.J.; Barbour, A.T.; Zaug, A.J.; Goodrich, K.J.; Mckay, A.E.; Wuttke, D.S.; Cech, T.R. The structure of human CST reveals a decameric assembly bound to telomeric DNA. Science 2020, 368, 1081–1085.
- Shastrula, P.K.; Rice, C.T.; Wang, Z.; Lieberman, P.M.; Skordalakes, E. Structural and functional analysis of an OB-fold in human Ctc1 implicated in telomere maintenance and bone marrow syndromes. Nucleic Acids Res. 2018, 46, 972–984.
- Chen, M.C.; Tippana, R.; Demeshkina, N.A.; Murat, P.; Balasubramanian, S.; Myong, S.; Ferre-D’Amare, A.R. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 2018, 558, 465–469.





