2. The Stressful Life of Leishmania Parasites
Leishmania parasites are the causative agent of the leishmaniases, a group of diverse diseases ranging in severity from a spontaneously healing skin ulcer to disfiguring mucocutaneous lesions or visceral disease, the latter often fatal if untreated
[3]. More than 12 million people suffer some form of the disease in tropical and subtropical regions of the world, including Central and South America, the Indian subcontinent, the Middle East, Central Asia, East Africa and the Mediterranean basin
[4].
This protist is transmitted to mammalians by insects; to complete its life cycle, therefore, the parasite requires adaptation to two different hosts. The promastigote stage (easily recognized by its flagellum) lives in the gut of phlebotomines (a.k.a., sand flies), mainly of the genera
Phlebotomus, Lutzomyia and
Psychodopygus. When the sand fly vector bites to feed on the mammalian host, promastigotes are inoculated into the dermis. Then, promastigotes are phagocytized by macrophages, but the parasites are able to survive inside the mature phagolysosome compartment, where they differentiate to the non-motile amastigote stage. Therefore, along its life cycle,
Leishmania faces dramatic changes in the environmental conditions, including temperature upshifts, acidic pH, oxidative stress and nutrient deprivation
[5]. Moreover, these changes are cyclical and reversible, when the parasite is transmitted back from the mammalian to the insect vector. Most of the environmental changes faced by the parasite might have a deleterious impact on structure and function of many
Leishmania proteins. In order to counteract these effects, cells have evolved the stress response, in which molecular chaperones are central components. As many molecular chaperones were first identified as being induced by heat shock, they are also known as heat shock proteins (HSPs) or in general stress proteins
[6]. In
Leishmania parasites, because of their stressful life cycle, a robust and versatile chaperone system was developed in order not only to act as cytoprotector against different stresses but also to tune stage differentiation and virulence development
[7].
3. The HSP70/HSP40 Chaperone System
Protein quality control is paramount for all molecular processes mediated by protein interactions that occur along the cell life. Together with protease systems and cellular mechanisms such as autophagy and lysosomal degradation, chaperones ensure that proteins are correctly folded and functional at the right place and time
[8]. Molecular chaperones are nanomachines specially engineered to interact with non-native conformations of proteins to avoid protein aggregation and assist protein folding. Usually, they bind to hydrophobic residues that are transiently exposed during initial folding but also present in damaged or denatured proteins; in their absence, protein aggregation may occur. The main classes of molecular chaperones are grouped in families, named according to the molecular weights of their prototypical members: small HPS (sHSPs), HSP40, HSP60, HSP70, HSP90 and HSP100
[8][9].
Among molecular chaperones, members of the HSP70 family are central hubs of many cellular processes
[10]. Because of this, it is not surprising that prototypical HSP70 (named DnaK in bacteria) is the most conserved protein present in all organisms
[11]. Moreover, HSP70 chaperones are key in protein quality control mechanisms. Thus, HSP70s interact with nascent polypeptides at the ribosome to assist de novo protein folding and with mis-folded or stress-denatured proteins; also, they are involved in the stabilization of partially unfolded proteins prior their translocation across membranes
[12][13][14].
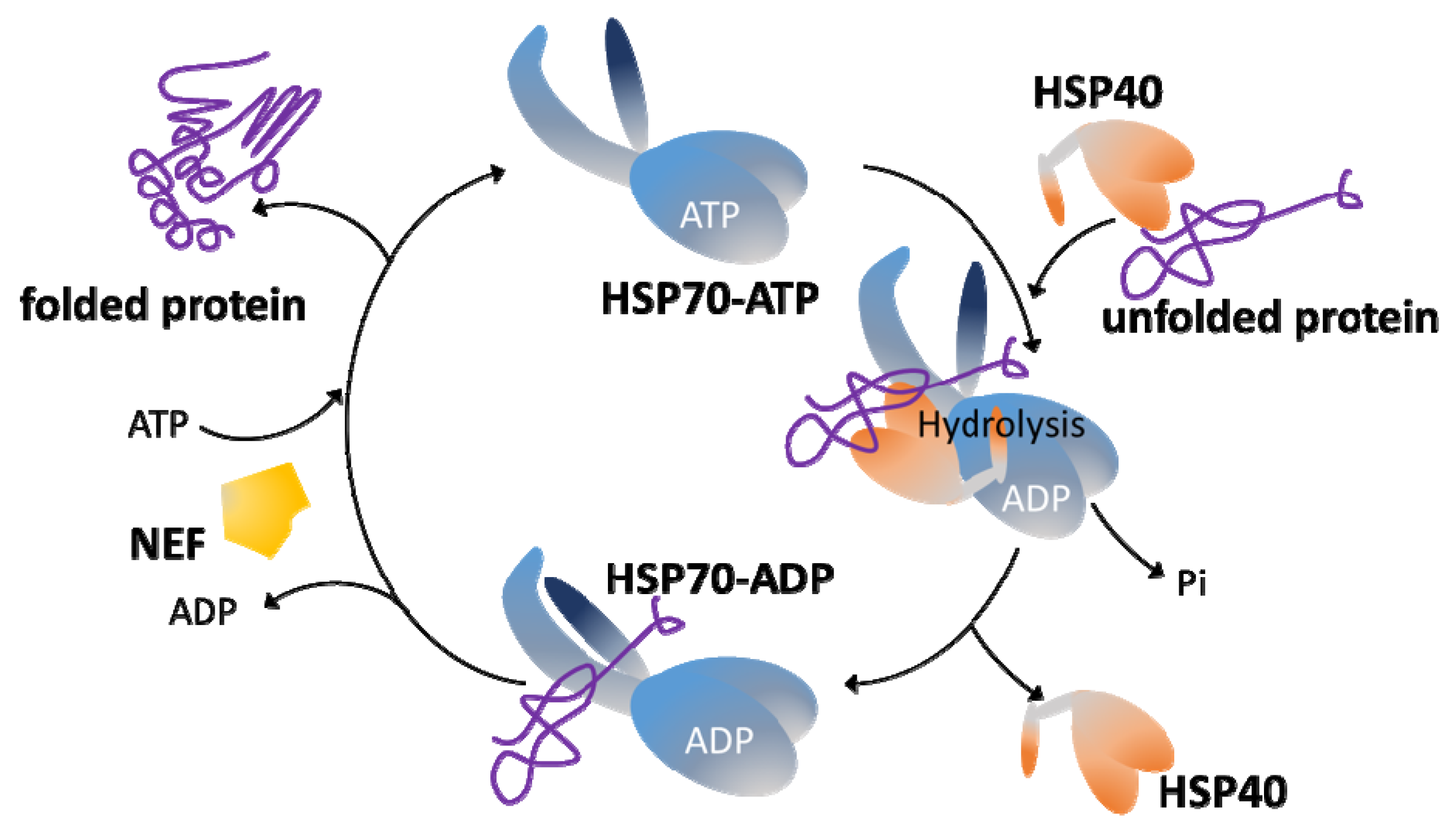
HSP70s have two functional domains, a substrate binding domain (SBD) and a nucleotide-binding domain (NBD). These domains work in a coordinate manner to accommodate the diverse functional roles played by HSP70s. Thus, the SBD moiety interacts with the target proteins, but this binding is transient, being modulated by an allosteric mechanism that involves cycles of ATP hydrolysis and ADP/ATP exchange in the NBD moiety (
Figure 1). Briefly, in the ATP-bound state, the HSP70 presents low affinity and fast exchange rates for the polypeptide substrates, while in the ADP-bound state, HSP70 has high affinity and low exchange rates for substrates
[10]. When ATP binds to the NBD, the entire HSP70 structure is affected, and the substrate-binding cleft of SBD opens to release the substrate (polypeptide). The subsequent ATP hydrolysis leads to a new conformational alteration (ADP-bound state) in the HSP70 structure that results in the trapping and tight binding of the target protein
[10]. Thus, binding and release alternating steps are important to stabilize non-native proteins and to promote their correct folding. Remarkably, HSP70s possess a weak intrinsic ATPase activity, but this is activated upon interaction with co-chaperones of the family HSP40/JDP. In fact, JDPs play a double role: they favor the binding of substrate proteins to HSP70 in its ATP-bound stage, but concomitantly a direct JDP-HSP70 interaction triggers the ATP hydrolysis and the transition to the ADP-bound state of HSP70, which has high affinity for the specific polypeptide brought over by a particular JDP. Another crucial player is the nucleotide exchange factor (NEF), which induces ADP dissociation and binding of a new ATP molecule, modifying HSP70 structure to the low-affinity state and consequently leading to substrate release
[15].
Figure 1. Overview of the HSP70/HSP40 chaperone system. An unfolded protein (the substrate) binds to an HSP40 member, and both form a complex with the HSP70 chaperone in its ATP-state, which has low affinity for polypeptides. The HSP40–HSP70 interaction triggers ATP hydrolysis and promotes a conformational change in the HSP70 (ADP-bound state) that results in a structure with high affinity for the protein substrate, which is bound into the substrate binding domain (SBD) of HSP70. Following substrate transfer, HSP40 leaves the complex, and the nucleotide exchange factor (NEF) is recruited to the HSP70–polypeptide complex, stimulating the ADP-by-ATP exchange. ATP binding induces both release of NEF and the folded polypeptide and leaves HSP70 ready for a new cycle.
4. J-Domain Proteins
HSP40s, also known as J-domain proteins (JDP), DNAJ-like or J proteins, are essential partners for HSP70 chaperones
[16]. These proteins are grouped because of the presence of a J-domain, whose prototype is that defined in
Escherichia coli DnaJ protein
[17]. JDPs can bind to substrate polypeptides by themselves, and their J-domain promotes ATP hydrolysis by the HSP70 protein, favoring the binding of polypeptides by the HSP70
[18]. Remarkably, but not surprisingly as they are responsible for substrate specificity, HSP40s, in a cell or into cellular compartments, outnumber HSP70 family members
[19]. Thus, 6 HSP40s have been found in
E. coli, 22 in
Saccharomyces cerevisiae, 41 in humans
[20], 49 in
Plasmodium falciparum [21] and 69 in
L. major [7]. In contrast, only three distinct DnaK genes exist in
E. coli; six distinct HSP70s are present in
P. falciparum, nine in
Leishmania and ten in humans
[7].
As mentioned above, the prototypical and founding member of this superfamily is the
E. coli DnaJ protein
[17], whose structure and functions have been elucidated in great detail
[22]. DnaJ contains four structural domains: an N-terminal J-domain, followed by a Gly/Phe (G-F)-rich domain, a Zn
2+-finger domain and a less-conserved C-terminal domain. The J-domain region is comprised of approximately 70 amino acids that fold into four α-helices (I–IV). The existence of a highly conserved His-Pro-Asp (HPD) tripeptide motif in the loop region between helices II and III is another structural feature of the J-domain; this motif is essential for the stimulation of HSP70 ATP hydrolysis
[23]. It is believed that J-domain only interacts with the ATP-bound HSP70 conformation at the interface between NBD and SBD moieties. Then, the HDP motif contacts key residues of the HSP70 ATP catalytic site, remodeling the NBD lobes to orientate the catalytic residues to a position optimal for ATP hydrolysis. Furthermore, the J-domain interacts with residues of the HSP70 SBD, promoting high affinity for the HSP70 ADP-bound state and efficient trapping of substrates
[10][22]. Moreover, many J-proteins directly bind substrates, favoring the specific interactions of HSP70 with particular polypeptides and linking in turn the HSP70 functions to particular cellular processes
[2]. Apart from the common J-domain, which is the distinctive feature of HSP40s, this class of proteins show a large structural divergence among them. Different HSP40s interact with a particular member of the HSP70 family; hence, HSP40s are contributing to the multi-functionality of the HSP70 machinery by specifying the target substrates and cellular processes in which the HSP70 chaperone activity is requested
[20][24].
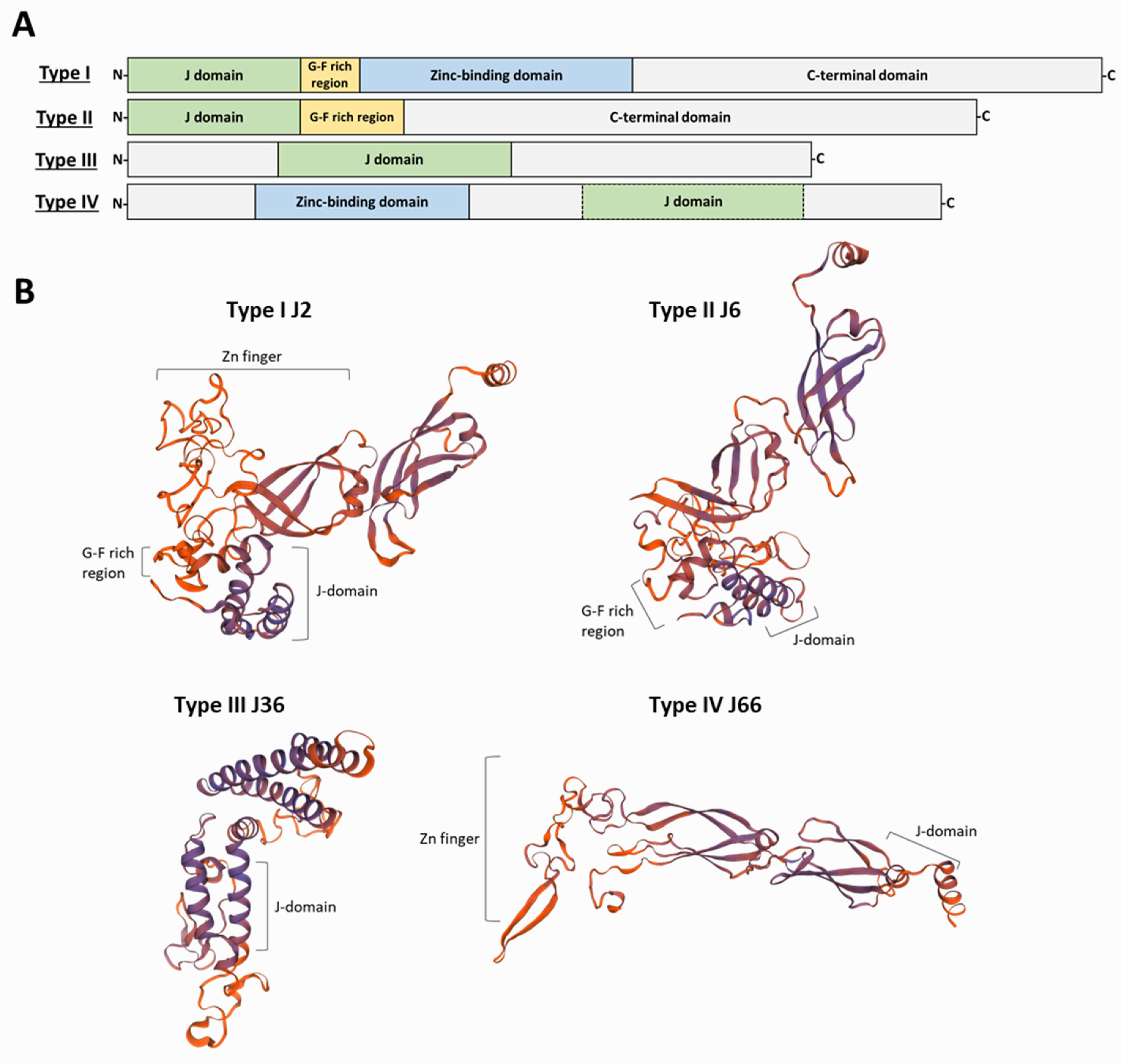
Based on the similarity to the domain architecture of the DnaJ, HSP40s have been grouped into four classes (
Figure 2). Type I proteins share the four characteristic domains of DnaJ: the N-terminal domain constituted by the J-domain; a glycine-phenylalanine (G-F)-rich linker segment; two β-sandwich C-terminal domains, which contain four repeats of the CxxCxGxG type (zinc finger-like region); and a dimerization domain involved in binding to the client proteins. Examples of proteins belonging to this class are the yeast Ydj1 and the human DnaJA1-4. Type II proteins share the J-domain, the G-F-rich linker and the C-terminal substrate binding domain but lack the zinc-finger domain; examples are the yeast Sis1 and the human DnaJB-1, -4 and -5
[24]. The G-F-rich region is also involved in determining the specificity of HSP40s for target proteins
[25]. Additionally, in class II JDPs, the G-F-rich region would be involved in an autoinhibitory mechanism, in which the G-F region initially blocks J-domain binding to HSP70
[15]. Type III proteins are heterogeneous in sequence and share only the J-domain with DnaJ; more often, they contain domains involved in specifying target interaction or sub-cellular localization
[20]. As mentioned above, within the J-domain, there exists the highly conserved HPD motif; however, for some J-domain containing proteins, a strict HPD sequence is not found. To denote this feature, Louw and coworkers proposed a fourth group (type IV) of HSP40s to include those proteins lacking the HPD sequence in their J-domains
[26]. Unlike type I and type II HSP40s, in which the J-domain always has an N-terminal location, in type III proteins the J-domain can be in any position along their sequence. It has been suggested that type I and type II HSP40s form complexes (dimers or tetramers) and are able to interact promiscuously with nascent polypeptides; also, they recognize mis-folded or aggregated proteins and cooperate with HSP70s in protein disaggregation
[10][27]. In contrast, type III HSP40s would have evolved to specifically interact with a limited number of HSP70 substrates or alternatively acting directly as a bait to locate an HSP70 to particular cellular places
[10][20]. Regarding type IV HSP40s, some authors have questioned whether they must be considered either members of the JDP family or only JDP-like proteins
[28]. Nevertheless, they should be considered to understand the evolutionary history of the family and its functional diversification. Moreover, for some HSP40s, the maintenance of their co-chaperone functions in the absence of a canonical J-domain has been reported
[29].
Figure 2. Classification of HSP40s into four types and representative
L. infantum proteins for each type. (
A) Type I proteins have in their structure the four typical domains, starting with a J-domain in the N-terminal end and a short G-F-rich region, followed by a zinc-binding domain that ends in the substrate-binding C-terminal domain. Type II proteins share the J-domain and the G-F-rich linker but lack the zinc-finger domain. Type III proteins show remarkable structure divergence and only share the J-domain, which is frequently located in the middle of the sequence. Type IV HSP40s include those proteins lacking the highly conserved HPD motif in their J-domains; the zinc-binding domain is absent in some type IV proteins. (
B) Protein structure of representative members of the HSP40 family in
L. infantum [30]. The J-domain consists of four α-helices. The G-F-rich linker is followed by two β-sandwich C-terminal domains which contain four repeats of the CxxCxGxG motif (zinc finger region) and a dimerization domain that is involved in binding to client polypeptides.