
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Cyril Favard | -- | 3057 | 2022-04-28 18:01:06 | | | |
| 2 | Beatrix Zheng | + 1 word(s) | 3058 | 2022-04-29 03:59:44 | | | | |
| 3 | Beatrix Zheng | Meta information modification | 3058 | 2022-04-29 04:01:42 | | |
Video Upload Options
The cell plasma membrane is mainly composed of phospholipids, cholesterol and embedded proteins, presenting a complex interface with the cell environment. Enveloped viruses are also surrounded by a lipidic membrane derived from the host-cell membrane and acquired during the assembly at and the budding from the host cell plasma membrane. In this perspective, model membranes, composed of selected lipid mixtures mimicking plasma membrane chemical and physical properties, are tools of choice to decipher the first steps of enveloped viruses assembly. Hereafter are detailled some of the existing artificial lipid membranes and their contribution in deciphering the assembly process of 3 well known envelopped virus, the human immunodeficiency virus 1 (HIV-1), the Influenza virus (IfV) and the Ebola virus (EboV).
1. Simplified Overview of Viral Assembly
Viral assembly, which is mainly driven by the self-assembly of viral structural proteins, precedes new particle release from the host cell. In the case of HIV-1, for example, 5 min are required for the viral particle to assemble, whereas budding and particle release occur, on average, 15 min later, independently of the cell type [1][2]. Viral assembly can be seen as a protein polymerization process involving three main steps, namely initiation, elongation and termination. In the case of enveloped virus assembly, initiation is always difficult to define. Here, the researchers define initiation as the nucleation step, i.e., the generation of a nucleus containing a small number of viral and host components, as follows: in the case of HIV-1, structural group-specific antigen (Gag) proteins, the viral RNA genome and host-cell membrane lipids; and, in the case of Influenza, the viral M1 and M2 proteins and the host-cell plasma membrane phosphophatidylserine. Initiation/nucleation requires an energetic barrier to be overcome [3]. From this perspective, the membrane can act as a dimensional catalyzer, which increases the probability of viral protein/viral protein and/or viral protein/genome complex encounters, as well as favoring assembly through entropic effects. In the very first step in membrane assembly, viral proteins are recruited at the membrane, generally via the interaction of the proteins with charged phospholipids.
2. Langmuir Monolayers: A Fine Tuning System to Monitor Membrane-Viral Proteins Interactions
3. Using Bicelles to Elucidate the Molecular Structures of Viral Proteins on Membranes
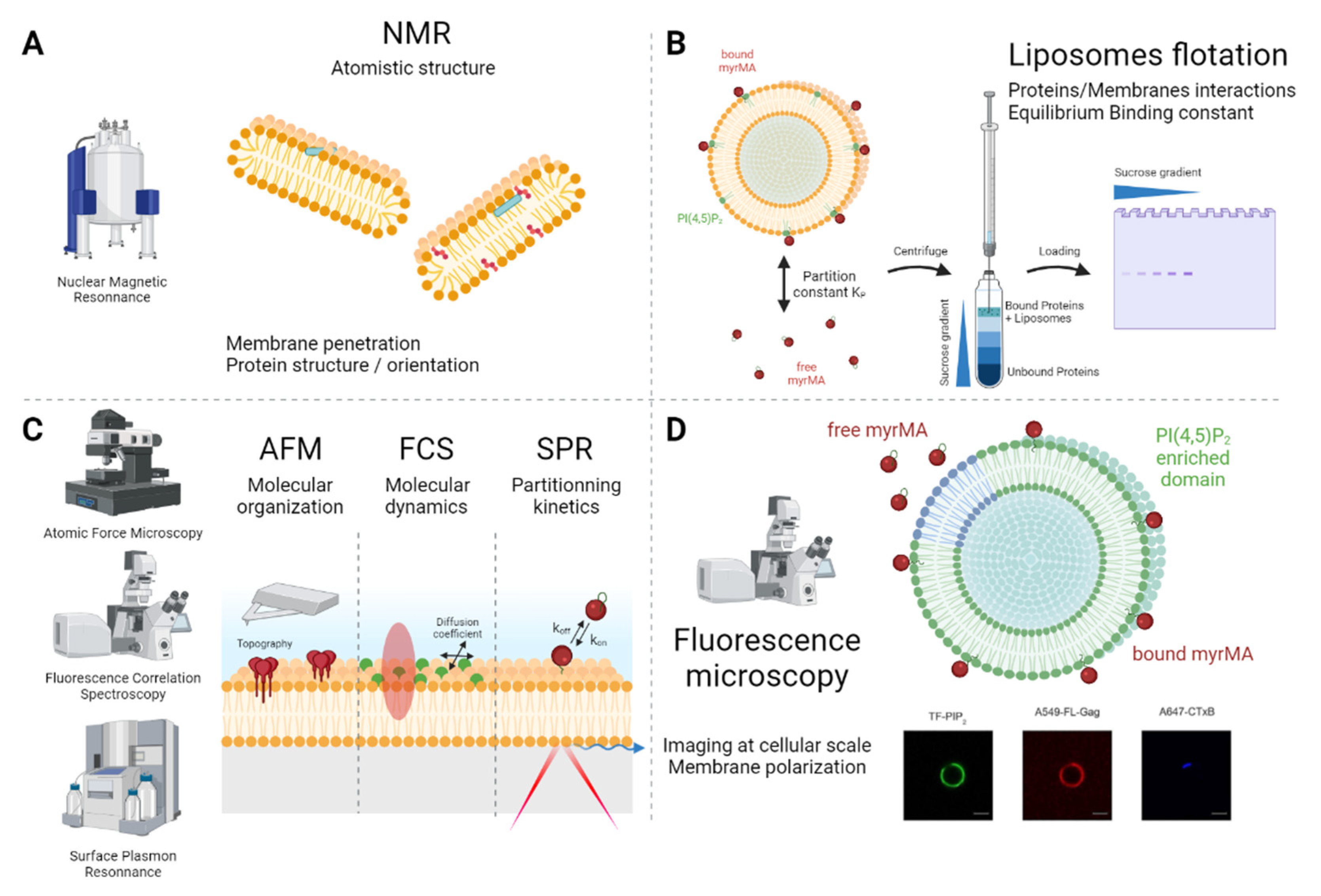
4. Unilamellar Vesicles
5. Supported and Tethered Bilayers: Planar Membranes
6. Giant Unilamellar Vesicles: The Closest Model to Cell Plasma Membranes
References
- Jouvenet, N.; Bieniasz, P.D.; Simon, S.M. Imaging the Biogenesis of Individual HIV-1 Virions in Live Cells. Nature 2008, 454, 236–240.
- Floderer, C.; Masson, J.-B.; Boilley, E.; Georgeault, S.; Merida, P.; El Beheiry, M.; Dahan, M.; Roingeard, P.; Sibarita, J.-B.; Favard, C.; et al. Single Molecule Localisation Microscopy Reveals How HIV-1 Gag Proteins Sense Membrane Virus Assembly Sites in Living Host CD4 T Cells. Sci. Rep. 2018, 8, 16283.
- Michaels, T.C.T.; Bellaiche, M.M.J.; Hagan, M.F.; Knowles, T.P.J. Kinetic Constraints on Self-Assembly into Closed Supramolecular Structures. Sci. Rep. 2017, 7, 12295.
- Malbec, M.; Sourisseau, M.; Guivel-Benhassine, F.; Porrot, F.; Blanchet, F.; Schwartz, O.; Casartelli, N. HIV-1 Nef Promotes the Localization of Gag to the Cell Membrane and Facilitates Viral Cell-to-Cell Transfer. Retrovirology 2013, 10, 80.
- Pirrone, G.F.; Emert-Sedlak, L.A.; Wales, T.E.; Smithgall, T.E.; Kent, M.S.; Engen, J.R. Membrane-Associated Conformation of HIV-1 Nef Investigated with Hydrogen Exchange Mass Spectrometry at a Langmuir Monolayer. Anal. Chem. 2015, 87, 7030–7035.
- Motsa, B.B.; Stahelin, R.V. Lipid–Protein Interactions in Virus Assembly and Budding from the Host Cell Plasma Membrane. Biochem. Soc. Trans. 2021, 49, 1633–1641.
- Adu-Gyamfi, E.; Soni, S.P.; Xue, Y.; Digman, M.A.; Gratton, E.; Stahelin, R.V. The Ebola Virus Matrix Protein Penetrates into the Plasma Membrane. J. Biol. Chem. 2013, 288, 5779–5789.
- Saad, J.S.; Miller, J.; Tai, J.; Kim, A.; Ghanam, R.H.; Summers, M.F. Structural Basis for Targeting HIV-1 Gag Proteins to the Plasma Membrane for Virus Assembly. Proc. Natl. Acad. Sci. USA 2006, 103, 11364–11369.
- Kerviel, A.; Thomas, A.; Chaloin, L.; Favard, C.; Muriaux, D. Virus Assembly and Plasma Membrane Domains: Which Came First? Virus Res. 2013, 171, 332–340.
- Vlach, J.; Saad, J.S. Trio Engagement via Plasma Membrane Phospholipids and the Myristoyl Moiety Governs HIV-1 Matrix Binding to Bilayers. Proc. Natl. Acad. Sci. USA 2013, 110, 3525–3530.
- Mercredi, P.Y.; Bucca, N.; Loeliger, B.; Gaines, C.R.; Mehta, M.; Bhargava, P.; Tedbury, P.R.; Charlier, L.; Floquet, N.; Muriaux, D.; et al. Structural and Molecular Determinants of Membrane Binding by the HIV-1 Matrix Protein. J. Mol. Biol. 2016, 428, 1637–1655.
- Charlier, L.; Louet, M.; Chaloin, L.; Fuchs, P.; Martinez, J.; Muriaux, D.; Favard, C.; Floquet, N. Coarse-Grained Simulations of the HIV-1 Matrix Protein Anchoring: Revisiting Its Assembly on Membrane Domains. Biophys. J. 2014, 106, 577–585.
- Wang, T.; Hong, M. Investigation of the Curvature Induction and Membrane Localization of the Influenza Virus M2 Protein Using Static and Off-Magic-Angle Spinning Solid-State Nuclear Magnetic Resonance of Oriented Bicelles. Biochemistry 2015, 54, 2214–2226.
- Rossman, J.S.; Lamb, R.A. Influenza Virus Assembly and Budding. Virology 2011, 411, 229–236.
- Zhou, W.; Resh, M.D. Differential Membrane Binding of the Human Immunodeficiency Virus Type 1 Matrix Protein. J. Virol. 1996, 70, 8540–8548.
- Dalton, A.K.; Ako-Adjei, D.; Murray, P.S.; Murray, D.; Vogt, V.M. Electrostatic Interactions Drive Membrane Association of the Human Immunodeficiency Virus Type 1 Gag MA Domain. J. Virol. 2007, 81, 6434–6445.
- Ehrlich, L.S.; Fong, S.; Scarlata, S.; Zybarth, G.; Carter, C. Partitioning of HIV-1 Gag and Gag-Related Proteins to Membranes. Biochemistry 1996, 35, 3933–3943.
- Yandrapalli, N.; Lubart, Q.; Tanwar, H.S.; Picart, C.; Mak, J.; Muriaux, D.; Favard, C. Self Assembly of HIV-1 Gag Protein on Lipid Membranes Generates PI(4,5)P2/Cholesterol Nanoclusters. Sci. Rep. 2016, 6, 39332.
- Dick, R.A.; Goh, S.L.; Feigenson, G.W.; Vogt, V.M. HIV-1 Gag Protein Can Sense the Cholesterol and Acyl Chain Environment in Model Membranes. Proc. Natl. Acad. Sci. USA 2012, 109, 18761–18766.
- Chukkapalli, V.; Hogue, I.B.; Boyko, V.; Hu, W.-S.; Ono, A. Interaction between the Human Immunodeficiency Virus Type 1 Gag Matrix Domain and Phosphatidylinositol-(4,5)-Bisphosphate Is Essential for Efficient Gag Membrane Binding. J. Virol. 2008, 82, 2405–2417.
- Bouamr, F.; Scarlata, S.; Carter, C. Role of Myristylation in HIV-1 Gag Assembly. Biochemistry 2003, 42, 6408–6417.
- Pérez Socas, L.B.; Ambroggio, E.E. The Influence of Myristoylation, Liposome Surface Charge and Nucleic Acid Interaction in the Partition Properties of HIV-1 Gag-N-Terminal Peptides to Membranes. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183421.
- Chukkapalli, V.; Oh, S.J.; Ono, A. Opposing Mechanisms Involving RNA and Lipids Regulate HIV-1 Gag Membrane Binding through the Highly Basic Region of the Matrix Domain. Proc. Natl. Acad. Sci. USA 2010, 107, 1600–1605.
- Sumner, C.; Kotani, O.; Liu, S.; Musier-Forsyth, K.; Sato, H.; Ono, A. Molecular Determinants in TRNA D-Arm Required for Inhibition of HIV-1 Gag Membrane Binding. J. Mol. Biol. 2022, 434, 167390.
- Todd, G.C.; Duchon, A.; Inlora, J.; Olson, E.D.; Musier-Forsyth, K.; Ono, A. Inhibition of HIV-1 Gag–Membrane Interactions by Specific RNAs. RNA 2017, 23, 395–405.
- Keller, H.; Kräusslich, H.-G.; Schwille, P. Multimerizable HIV Gag Derivative Binds to the Liquid-Disordered Phase in Model Membranes. Cell. Microbiol. 2013, 15, 237–247.
- Urbančič, I.; Brun, J.; Shrestha, D.; Waithe, D.; Eggeling, C.; Chojnacki, J. Lipid Composition but Not Curvature Is the Determinant Factor for the Low Molecular Mobility Observed on the Membrane of Virus-Like Vesicles. Viruses 2018, 10, 415.
- Gregoriades, A. Interaction of Influenza M Protein with Viral Lipid and Phosphatidylcholine Vesicles. J. Virol. 1980, 36, 470–479.
- Oxford, J.S.; Hockley, D.J.; Heath, T.D.; Patterson, S. The Interaction of Influenza Virus Haemagglutinin with Phospholipid Vesicles—Morphological and Immunological Studies. J. Gen. Virol. 1981, 52, 329–343.
- Kerviel, A.; Dash, S.; Moncorgé, O.; Panthu, B.; Prchal, J.; Décimo, D.; Ohlmann, T.; Lina, B.; Favard, C.; Decroly, E.; et al. Involvement of an Arginine Triplet in M1 Matrix Protein Interaction with Membranes and in M1 Recruitment into Virus-Like Particles of the Influenza A(H1N1)Pdm09 Virus. PLoS ONE 2016, 11, e0165421.
- Baudin, F.; Petit, I.; Weissenhorn, W.; Ruigrok, R.W. In Vitro Dissection of the Membrane and RNP Binding Activities of Influenza Virus M1 Protein. Virology 2001, 281, 102–108.
- Ruigrok, R.W.; Barge, A.; Durrer, P.; Brunner, J.; Ma, K.; Whittaker, G.R. Membrane Interaction of Influenza Virus M1 Protein. Virology 2000, 267, 289–298.
- Kordyukova, L.V.; Konarev, P.V.; Fedorova, N.V.; Shtykova, E.V.; Ksenofontov, A.L.; Loshkarev, N.A.; Dadinova, L.A.; Timofeeva, T.A.; Abramchuk, S.S.; Moiseenko, A.V.; et al. The Cytoplasmic Tail of Influenza A Virus Hemagglutinin and Membrane Lipid Composition Change the Mode of M1 Protein Association with the Lipid Bilayer. Membranes 2021, 11, 772.
- Huang, R.T.C.; Warn, K.; Klenk, H.-D.; Rott, R. Association of the Envelope Glycoproteins of Influenza Virus with Liposomes—a Model Study on Viral Envelope Assembly. Virology 1979, 97, 212–217.
- Bailey, A.; Zhukovsky, M.; Gliozzi, A.; Chernomordik, L.V. Liposome Composition Effects on Lipid Mixing between Cells Expressing Influenza Virus Hemagglutinin and Bound Liposomes. Arch. Biochem. Biophys. 2005, 439, 211–221.
- Rossman, J.S.; Jing, X.; Leser, G.P.; Lamb, R.A. Influenza Virus M2 Protein Mediates ESCRT-Independent Membrane Scission. Cell 2010, 142, 902–913.
- Martyna, A.; Gómez-Llobregat, J.; Lindén, M.; Rossman, J.S. Curvature Sensing by a Viral Scission Protein. Biochemistry 2016, 55, 3493–3496.
- Martyna, A.; Bahsoun, B.; Madsen, J.J.; Jackson, F.S.J.S.; Badham, M.D.; Voth, G.A.; Rossman, J.S. Cholesterol Alters the Orientation and Activity of the Influenza Virus M2 Amphipathic Helix in the Membrane. J. Phys. Chem. B 2020, 124, 6738–6747.
- Ruigrok, R.W.H.; Schoehn, G.; Dessen, A.; Forest, E.; Volchkov, V.; Dolnik, O.; Klenk, H.-D.; Weissenhorn, W. Structural Characterization and Membrane Binding Properties of the Matrix Protein VP40 of Ebola Virus11Edited by J. Karn. J. Mol. Biol. 2000, 300, 103–112.
- Scianimanico, S. Membrane Association Induces a Conformational Change in the Ebola Virus Matrix Protein. EMBO J. 2000, 19, 6732–6741.
- Johnson, K.A.; Taghon, G.J.F.; Scott, J.L.; Stahelin, R.V. The Ebola Virus Matrix Protein, VP40, Requires Phosphatidylinositol 4,5-Bisphosphate (PI(4,5)P2) for Extensive Oligomerization at the Plasma Membrane and Viral Egress. Sci. Rep. 2016, 6, 19125.
- Valbuena, A.; Maity, S.; Mateu, M.G.; Roos, W.H. Visualization of Single Molecules Building a Viral Capsid Protein Lattice through Stochastic Pathways. ACS Nano 2020, 14, 8724–8734.
- Favard, C.; Chojnacki, J.; Merida, P.; Yandrapalli, N.; Mak, J.; Eggeling, C.; Muriaux, D. HIV-1 Gag Specifically Restricts PI(4,5)P2 and Cholesterol Mobility in Living Cells Creating a Nanodomain Platform for Virus Assembly. Sci. Adv. 2019, 5, eaaw8651.
- Miles, P.; Cassidy, P.; Donlon, L.; Yarkoni, O.; Frankel, D. In Vitro Assembly of a Viral Envelope. Soft Matter 2015, 11, 7722–7727.
- Hilsch, M.; Goldenbogen, B.; Sieben, C.; Höfer, C.T.; Rabe, J.P.; Klipp, E.; Herrmann, A.; Chiantia, S. Influenza A Matrix Protein M1 Multimerizes upon Binding to Lipid Membranes. Biophys. J. 2014, 107, 912–923.
- Bobone, S.; Hilsch, M.; Storm, J.; Dunsing, V.; Herrmann, A.; Chiantia, S. Phosphatidylserine Lateral Organization Influences the Interaction of Influenza Virus Matrix Protein 1 with Lipid Membranes. J. Virol. 2017, 91, e00267-17.
- Höfer, C.T.; Di Lella, S.; Dahmani, I.; Jungnick, N.; Bordag, N.; Bobone, S.; Huang, Q.; Keller, S.; Herrmann, A.; Chiantia, S. Structural Determinants of the Interaction between Influenza A Virus Matrix Protein M1 and Lipid Membranes. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1123–1134.
- Cremer, P.S.; Boxer, S.G. Formation and Spreading of Lipid Bilayers on Planar Glass Supports. J. Phys. Chem. B 1999, 103, 2554–2559.
- Wen, Y.; Feigenson, G.W.; Vogt, V.M.; Dick, R.A. Mechanisms of PI(4,5)P2 Enrichment in HIV-1 Viral Membranes. J. Mol. Biol. 2020, 432, 5343–5364.
- Gui, D.; Gupta, S.; Xu, J.; Zandi, R.; Gill, S.; Huang, I.-C.; Rao, A.L.N.; Mohideen, U. A Novel Minimal in Vitro System for Analyzing HIV-1 Gag-Mediated Budding. J. Biol. Phys. 2015, 41, 135–149.
- Van Engelenburg, S.B.; Shtengel, G.; Sengupta, P.; Waki, K.; Jarnik, M.; Ablan, S.D.; Freed, E.O.; Hess, H.F.; Lippincott-Schwartz, J. Distribution of ESCRT Machinery at HIV Assembly Sites Reveals Virus Scaffolding of ESCRT Subunits. Science 2014, 343, 653–656.
- Carlson, L.-A.; Hurley, J.H. In Vitro Reconstitution of the Ordered Assembly of the Endosomal Sorting Complex Required for Transport at Membrane-Bound HIV-1 Gag Clusters. Proc. Natl. Acad. Sci. USA 2012, 109, 16928–16933.
- Dahmani, I.; Ludwig, K.; Chiantia, S. Influenza A Matrix Protein M1 Induces Lipid Membrane Deformation via Protein Multimerization. Biosci. Rep. 2019, 39, BSR20191024.
- Saletti, D.; Radzimanowski, J.; Effantin, G.; Midtvedt, D.; Mangenot, S.; Weissenhorn, W.; Bassereau, P.; Bally, M. The Matrix Protein M1 from Influenza C Virus Induces Tubular Membrane Invaginations in an in Vitro Cell Membrane Model. Sci. Rep. 2017, 7, 40801.
- Soni, S.P.; Adu-Gyamfi, E.; Yong, S.S.; Jee, C.S.; Stahelin, R.V. The Ebola Virus Matrix Protein Deeply Penetrates the Plasma Membrane: An Important Step in Viral Egress. Biophys. J. 2013, 104, 1940–1949.
- Soni, S.P.; Stahelin, R.V. The Ebola Virus Matrix Protein VP40 Selectively Induces Vesiculation from Phosphatidylserine-Enriched Membranes. J. Biol. Chem. 2014, 289, 33590–33597.




