
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Cassandra Terry | -- | 7369 | 2022-04-28 11:14:17 | | | |
| 2 | Camila Xu | -3977 word(s) | 3392 | 2022-04-29 02:37:26 | | |
Video Upload Options
Cases of Type 2 Diabetes Mellitus (T2DM) are increasing at an alarming rate due to the rise in obesity, sedentary lifestyles, glucose-rich diets and other factors. Numerous studies have increasingly illustrated the pivotal role that human islet amyloid polypeptide (hIAPP) plays in the pathology of T2DM through damage and subsequent loss of pancreatic β-cell mass. Here researchers provide an up-to-date summary of recent progress in the field, highlighting factors that contribute to hIAPP misfolding and aggregation which have been linked to β-cell cytotoxicity. Understanding the structure of hIAPP and how these factors affect amyloid formation will help better understand how hIAPP misfolds and aggregates and, importantly, help identify potential therapeutic targets for inhibiting amyloidosis so alternate and more effective treatments for T2DM can be developed.
1. Introduction
2. Main Text
2.1. The Human Islet Amyloid Polypeptide and Its Link to Diabetes
3. The Structure of hIAPP
4. Important Residues Involved in hIAPP Aggregation
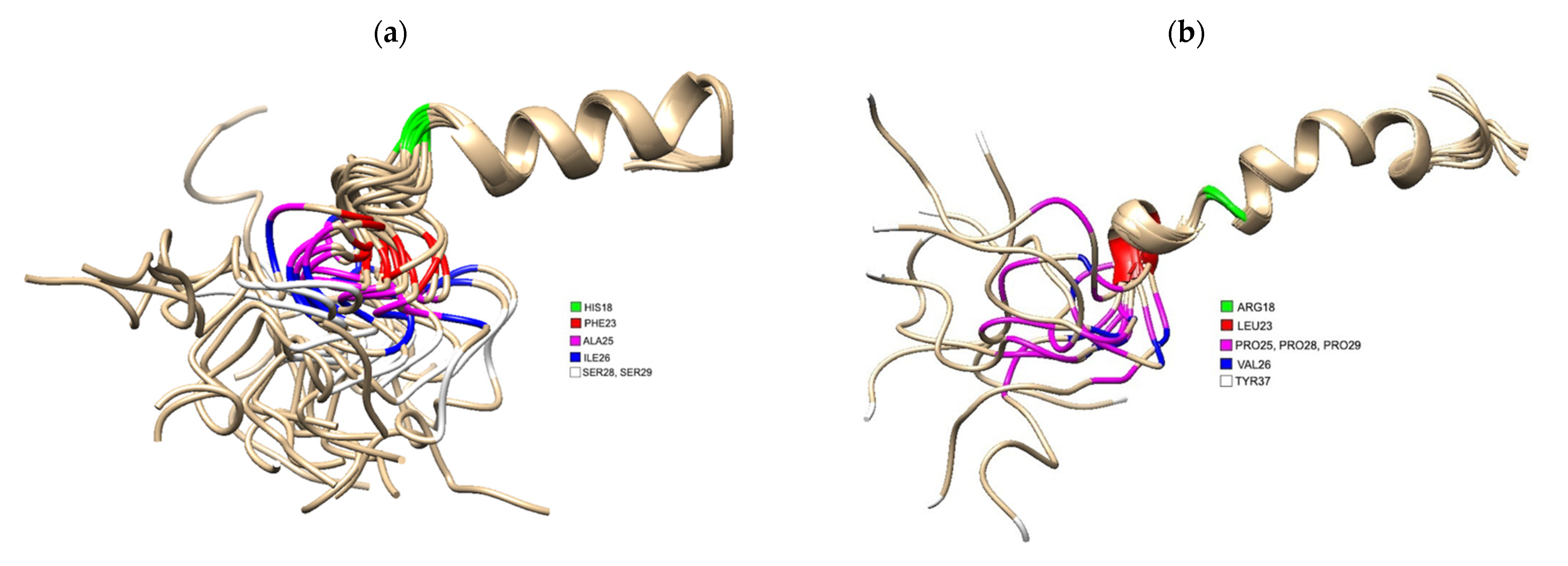 Figure 1. (a) 3D Structure of human IAPP residues 1–37 of the mature protein, solved by NMR (PDB 5MGQ) with key residues highlighted to show the difference compared to rat IAPP. The C and N terminus are located on the left and right, respectively. Residues were highlighted using USCF Chimera. HIS18 is highlighted in green, PHE23 is red, ALA25 is magenta, ILE26 is blue and SER28 and SER 29 are white.; (b) 3D Structure of rat IAPP residues 1–37, solved by NMR (PDB 2KJ7). Residues differing from hIAPP were highlighted using USCF Chimera. ARG18 is highlighted in green, LEU23 is red, PRO25, PRO28 and PRO 29 are magenta, VAL26 and TYR37 is white.
Figure 1. (a) 3D Structure of human IAPP residues 1–37 of the mature protein, solved by NMR (PDB 5MGQ) with key residues highlighted to show the difference compared to rat IAPP. The C and N terminus are located on the left and right, respectively. Residues were highlighted using USCF Chimera. HIS18 is highlighted in green, PHE23 is red, ALA25 is magenta, ILE26 is blue and SER28 and SER 29 are white.; (b) 3D Structure of rat IAPP residues 1–37, solved by NMR (PDB 2KJ7). Residues differing from hIAPP were highlighted using USCF Chimera. ARG18 is highlighted in green, LEU23 is red, PRO25, PRO28 and PRO 29 are magenta, VAL26 and TYR37 is white.5. The Role of hIAPP Oligomeric Intermediates in T2DM
6. Mechanisms of hIAPP Cytotoxicity
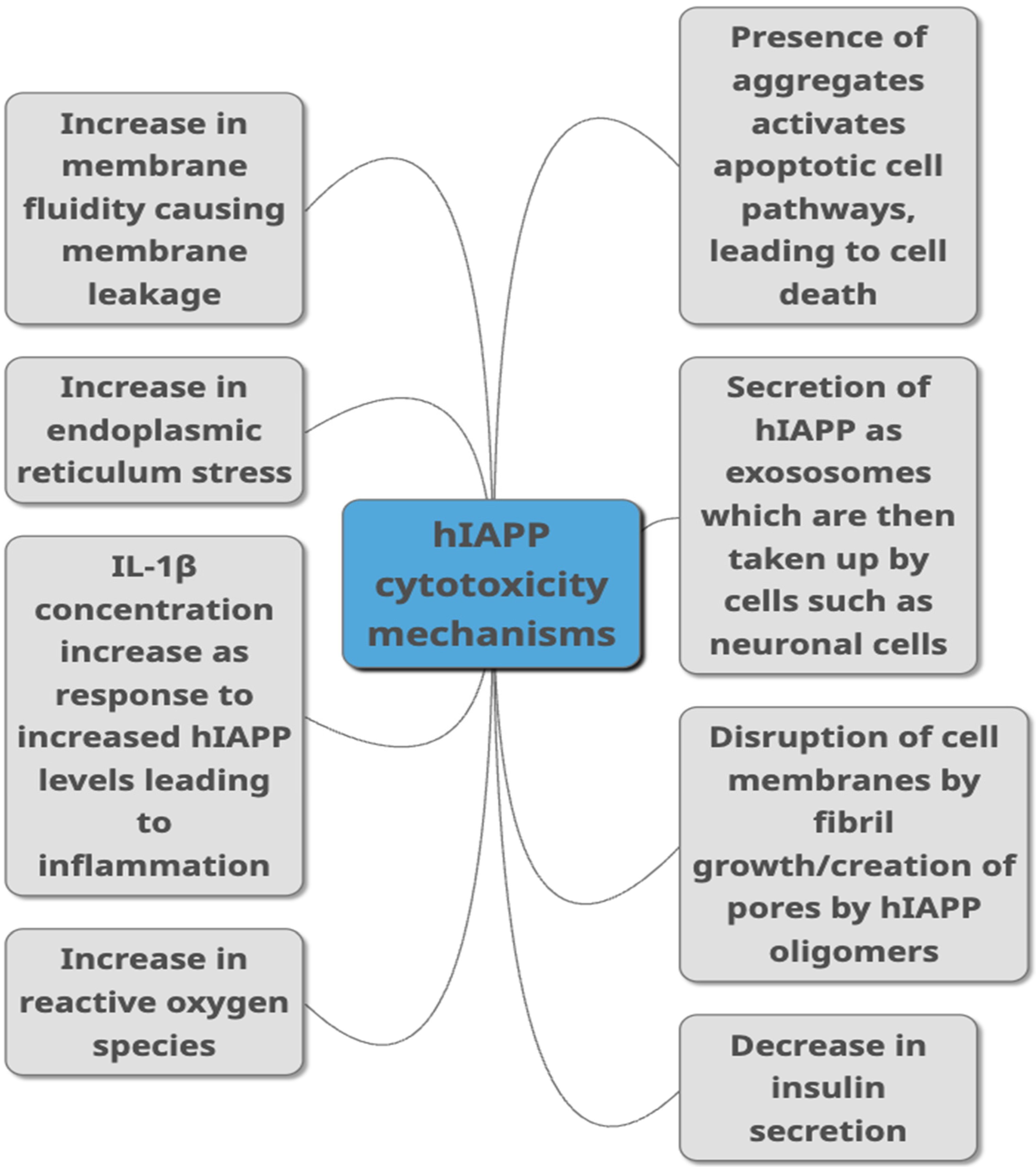 Figure 2. Proposed mechanisms through which hIAPP is linked to toxicity in T2DM as summarised from studies published in peer-reviewed journals. They include different ways of damaging the cells such as causing damage to the membrane, causing cell inflammation, damaging cells through aggregate build-up, by increasing ROS production and ER stress. Decreased insulin secretion, activation of apoptopic pathways and secretion via exosomes are other pathways suggested to link hIAPP with cytotoxicity.
Figure 2. Proposed mechanisms through which hIAPP is linked to toxicity in T2DM as summarised from studies published in peer-reviewed journals. They include different ways of damaging the cells such as causing damage to the membrane, causing cell inflammation, damaging cells through aggregate build-up, by increasing ROS production and ER stress. Decreased insulin secretion, activation of apoptopic pathways and secretion via exosomes are other pathways suggested to link hIAPP with cytotoxicity.7. Acceleration of hIAPP Misfolding and Aggregation
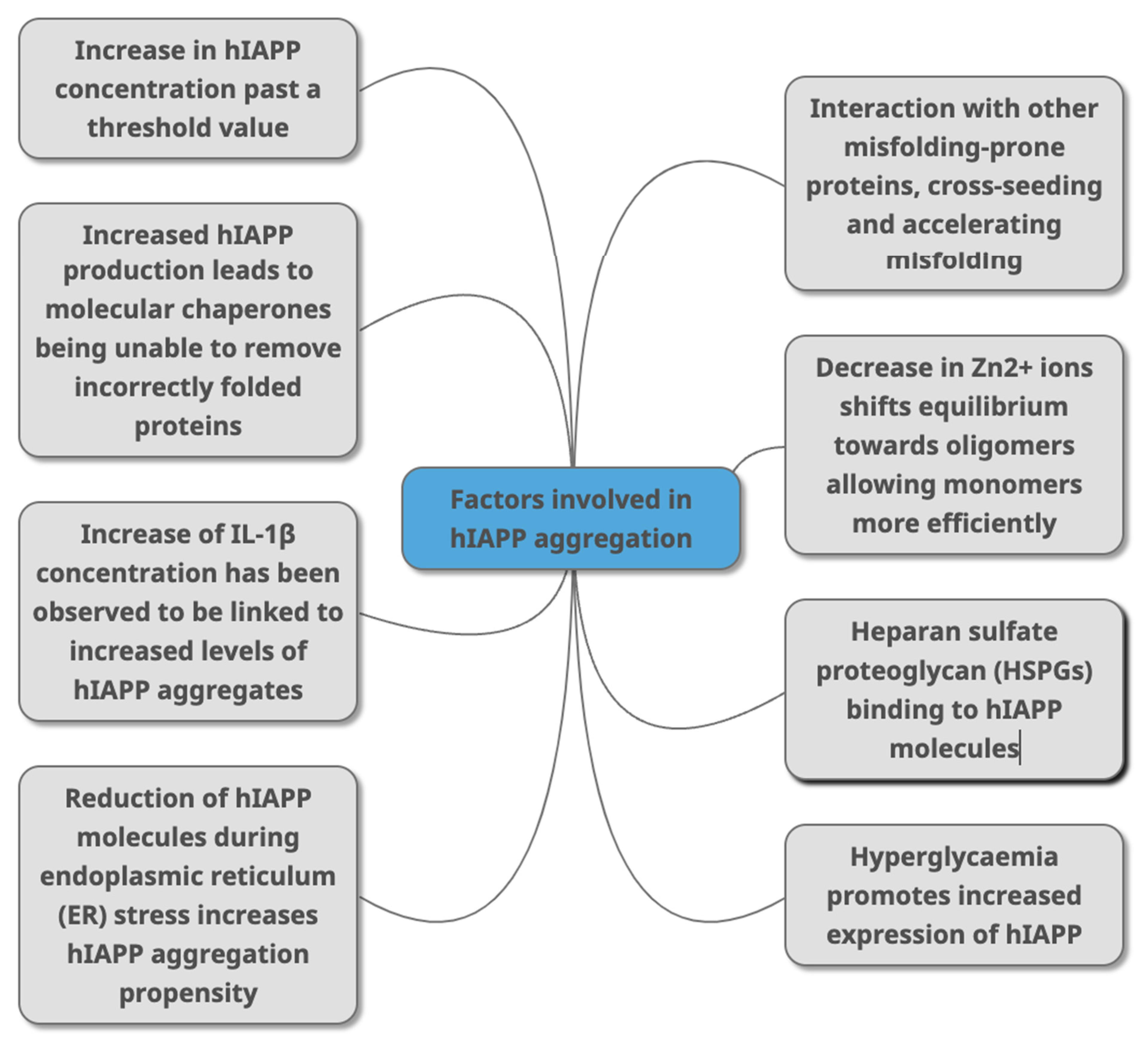 Figure 3. Factors that have been identified in peer-reviewed journals as contributing to hIAPP aggregation. This can occur due to change of concentrations of different molecules (HSPGs, Zn2+ IL-1β, Aβ, alpha synuclein) that directly or indirectly interact with hIAPP as part of various metabolic mechanisms (molecular chaperones, and through conditions linked with T2DM (hyperglycaemia, ER stress).
Figure 3. Factors that have been identified in peer-reviewed journals as contributing to hIAPP aggregation. This can occur due to change of concentrations of different molecules (HSPGs, Zn2+ IL-1β, Aβ, alpha synuclein) that directly or indirectly interact with hIAPP as part of various metabolic mechanisms (molecular chaperones, and through conditions linked with T2DM (hyperglycaemia, ER stress).References
- Mittal, K.; Mani, R.; Katare, D. Type 3 Diabetes: Cross Talk between Differentially Regulated Proteins of Type 2 Diabetes Mellitus and Alzheimer’s Disease. Sci. Rep. 2016, 6, 25589.
- Steck, A.; Winter, W. Review on monogenic diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 2011, 18, 252–258.
- Tan, S.; Mei Wong, J.; Sim, Y.; Wong, S.; Mohamed Elhassan, S.; Tan, S.; Ling Lim, G.; Rong Tay, N.; Annan, N.; Bhattamisra, S.; et al. Type 1 and 2 diabetes mellitus: A review on current treatment approach and gene therapy as potential intervention. Diabetes Metab. Syndr. Clin. Res. Rev. 2019, 13, 364–372.
- Mack, L.; Tomich, P. Gestational Diabetes. Obstet. Gynecol. Clin. N. Am. 2017, 44, 207–217.
- Nguyen, T.; Ta, Q.; Nguyen, T.; Nguyen, T.; Van Giau, V. Type 3 diabetes and its role implications in Alzheimer’s disease. Int. J. Mol. Sci. 2020, 21, 3165.
- Giugliano, D.; Ceriello, A.; Esposito, K. Glucose metabolism and hyperglycemia. Am. J. Clin. Nutr. 2008, 87, 217S–222S.
- Stratton, I. Association of glycaemia with macrovascular and microvascular complications of type 2 diabetes (UKPDS 35): Prospective observational study. BMJ 2000, 321, 405–412.
- World Health Organization. Available online: https://www.who.int (accessed on 6 April 2022).
- Ling, C.; Rönn, T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019, 29, 1028–1044.
- Xue, A.; Wu, Y.; Zhu, Z.; Zhang, F.; Kemper, K.E.; Zheng, Z.; Yengo, L.; Lloyd-Jones, L.R.; Sidorenko, J.; Wu, Y.; et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat. Commun. 2018, 9, 2941.
- Vujkovic, M.; Keaton, J.M.; Lynch, J.A.; Miller, D.R.; Zhou, J.; Tcheandjieu, C.; Huffman, J.E.; Assimes, T.L.; Lorenz, K.; Zhu, X.; et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 2020, 52, 680–691.
- Chooi, Y.C.; Ding, C.; Magkos, F. The epidemiology of obesity. Metabolism 2019, 92, 6–10.
- Toplak, H.; Leitner, D.; Harreiter, J.; Hoppichler, F.; Wascher, T.; Schindler, K.; Ludvik, B. “Diabesity”—Adipositas und Typ-2-Diabetes (Update 2019). Wien. Klin. Wochenschr. 2019, 131, 71–76.
- Pulgaron, E.R.; Delamater, A.M. Obesity and Type 2 Diabetes in Children: Epidemiology and Treatment. Curr. Diabetes Rep. 2014, 14, 508.
- Weickert, M.O.; Pfeiffer, A.F. Impact of Dietary Fiber Consumption on Insulin Resistance and the Prevention of Type 2 Diabetes. J. Nutr. 2018, 148, 7–12.
- Hill, J.; Nielsen, M.; Fox, M.H. Understanding the Social Factors That Contribute to Diabetes: A Means to Informing Health Care and Social Policies for the Chronically Ill. Perm. J. 2013, 17, 67–72.
- Li, W.-Z.; Stirling, K.; Yang, J.-J.; Zhang, L. Gut microbiota and diabetes: From correlation to causality and mechanism. World J. Diabetes 2020, 11, 293–308.
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590.
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772.
- Foretz, M.; Guigas, B.; Viollet, B. Understanding the glucoregulatory mechanisms of metformin in type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2019, 15, 569–589.
- Sola, D.; Rossi, L.; Schianca, G.P.C.; Maffioli, P.; Bigliocca, M.; Mella, R.; Corlianò, F.; Fra, G.P.; Bartoli, E.; Derosa, G. State of the art paper Sulfonylureas and their use in clinical practice. Arch. Med. Sci. 2015, 4, 840–848.
- Bishoyi, A.K.; Roham, P.H.; Rachineni, K.; Save, S.; Hazari, M.A.; Sharma, S.; Kumar, A. Human islet amyloid polypeptide (hIAPP)—A curse in type II diabetes mellitus: Insights from structure and toxicity studies. Biol. Chem. 2021, 402, 133–153.
- Pandey, A.; Chawla, S.; Guchhait, P. Type-2 diabetes: Current understanding and future perspectives. IUBMB Life 2015, 67, 506–513.
- Hanabusa, T.; Kubo, K.; Oki, C.; Nakano, Y.; Okai, K.; Sanke, T.; Nanjo, K. Islet amyloid polypeptide (IAPP) secretion from islet cells and its plasma concentration in patients with non-insulin-dependent diabetes mellitus. Diabetes Res. Clin. Pract. 1992, 15, 89–96.
- Scollo, F.; La Rosa, C. Amyloidogenic Intrinsically Disordered Proteins: New Insights into Their Self-Assembly and Their Interaction with Membranes. Life 2020, 10, 144.
- Castillo, M.J.; Scheen, A.J.; Lefèbvre, P.J. Amylin/islet amyloid polypeptide: Biochemistry, physiology, patho-physiology. Diabete Metab. 1995, 21, 3–25.
- Westermark, P.; Engström, U.; Westermark, G.T.; Johnson, K.H.; Permerth, J.; Betsholtz, C. Islet amyloid polypeptide (IAPP) and pro-IAPP immunoreactivity in human islets of Langerhans. Diabetes Res. Clin. Pract. 1989, 7, 219–226.
- Akter, R.; Cao, P.; Noor, H.; Ridgway, Z.; Tu, L.-H.; Wang, H.; Wong, A.G.; Zhang, X.; Abedini, A.; Schmidt, A.M.; et al. Islet Amyloid Polypeptide: Structure, Function, and Pathophysiology. J. Diabetes Res. 2016, 2016, 2798269.
- Wang, J.; Xu, J.; Finnerty, J.; Furuta, M.; Steiner, D.F.; Verchere, C.B. The Prohormone Convertase Enzyme 2 (PC2) Is Essential for Processing Pro-Islet Amyloid Polypeptide at the NH2-Terminal Cleavage Site. Diabetes 2001, 50, 534–539.
- Westermark, P.; Andersson, A.; Westermark, G.T. Islet Amyloid Polypeptide, Islet Amyloid, and Diabetes Mellitus. Physiol. Rev. 2011, 91, 795–826.
- Cao, P.; Abedini, A.; Raleigh, D.P. Aggregation of islet amyloid polypeptide: From physical chemistry to cell biology. Curr. Opin. Struct. Biol. 2012, 23, 82–89.
- Mulder, H.; Ahren, B.; Sundler, F. Islet amyloid polypeptide and insulin gene expression are regulated in parallel by glucose in vivo in rats. Am. J. Physiol. Metab. 1996, 271, E1008–E1014.
- Mulder, H.; Gebre-Medhin, S.; Betsholtz, C.; Sundler, F.; Ahrén, B. Islet amyloid polypeptide (amylin)-deficient mice develop a more severe form of alloxan-induced diabetes. Am. J. Physiol. Endocrinol. Metab. 2000, 278, E684–E691.
- Moore, S.J.; Sonar, K.; Bharadwaj, P.; Deplazes, E.; Mancera, R.L. Characterisation of the Structure and Oligomerisation of Islet Amyloid Polypeptides (IAPP): A Review of Molecular Dynamics Simulation Studies. Molecules 2018, 23, 2142.
- Higham, C.E.; Jaikaran, E.T.; Fraser, P.E.; Gross, M.; Clark, A. Preparation of synthetic human islet amyloid polypeptide (IAPP) in a stable conformation to enable study of conversion to amyloid-like fibrils. FEBS Lett. 2000, 470, 55–60.
- Bedrood, S.; Li, Y.; Isas, J.M.; Hegde, B.G.; Baxa, U.; Haworth, I.S.; Langen, R. Fibril Structure of Human Islet Amyloid Polypeptide. J. Biol. Chem. 2012, 287, 5235–5241.
- Qiao, Q.; Bowman, G.R.; Huang, X. Dynamics of an Intrinsically Disordered Protein Reveal Metastable Conformations That Potentially Seed Aggregation. J. Am. Chem. Soc. 2013, 135, 16092–16101.
- Fu, Z.; Aucoin, D.; Davis, J.; Van Nostrand, W.E.; Smith, S.O. Mechanism of Nucleated Conformational Conversion of Aβ42. Biochemistry 2015, 54, 4197–4207.
- Paul, F.; Weikl, T.R. How to Distinguish Conformational Selection and Induced Fit Based on Chemical Relaxation Rates. PLOS Comput. Biol. 2016, 12, e1005067.
- Cao, Q.; Boyer, D.; Sawaya, M.; Ge, P.; Eisenberg, D. Cryo-EM structure and inhibitor design of humanIAPP (amylin) fibrils. Nat. Struct. Mol. Biol. 2020, 27, 653–659.
- Gallardo, R.; Iadanza, M.G.; Xu, Y.; Heath, G.R.; Foster, R.; Radford, S.E.; Ranson, N.A. Fibril structures of diabetes-related amylin variants reveal a basis for surface-templated assembly. Nat. Struct. Mol. Biol. 2020, 27, 1048–1056.
- Röder, C.; Kupreichyk, T.; Gremer, L.; Schäfer, L.U.; Pothula, K.R.; Ravelli, R.B.G.; Willbold, D.; Hoyer, W.; Schröder, G.F. Cryo-EM structure of islet amyloid polypeptide fibrils reveals similarities with amyloid-β fibrils. Nat. Struct. Mol. Biol. 2020, 27, 660–667.
- Zhang, X.; Li, D.; Zhu, X.; Wang, Y.; Zhu, P. Structural characterization and cryo-electron tomography analysis of human islet amyloid polypeptide suggest a synchronous process of the hIAPP1−37 amyloid fibrillation. Biochem. Biophys. Res. Commun. 2020, 533, 125–131.
- Cao, Q.; Boyer, D.R.; Sawaya, M.R.; Abskharon, R.; Saelices, L.; Nguyen, B.A.; Lu, J.; Murray, K.A.; Kandeel, F.; Eisenberg, D.S. Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores. Nat. Struct. Mol. Biol. 2021, 28, 724–730.
- Li, M.S.; Klimov, D.K.; Straub, J.E.; Thirumalai, D. Probing the mechanisms of fibril formation using lattice models. J. Chem. Phys. 2008, 129, 175101.
- Brender, J.; Dürr, U.H.; Heyl, D.; Budarapu, M.B.; Ramamoorthy, A. Membrane fragmentation by an amyloidogenic fragment of human Islet Amyloid Polypeptide detected by solid-state NMR spectroscopy of membrane nanotubes. Biochim. Biophys. Acta (BBA) 2007, 1768, 2026–2029.
- Apostolidou, M.; Jayasinghe, S.A.; Langen, R. Structure of α-Helical Membrane-bound Human Islet Amyloid Polypeptide and Its Implications for Membrane-mediated Misfolding. J. Biol. Chem. 2008, 283, 17205–17210.
- Zhang, X.; Clair, J.R.S.; London, E.; Raleigh, D.P. Islet Amyloid Polypeptide Membrane Interactions: Effects of Membrane Composition. Biochemistry 2017, 56, 376–390.
- Sasahara, K. Membrane-mediated amyloid deposition of human islet amyloid polypeptide. Biophys. Rev. 2018, 10, 453–462.
- Caillon, L.; Hoffmann, A.R.F.; Botz, A.; Khemtemourian, L. Molecular Structure, Membrane Interactions, and Toxicity of the Islet Amyloid Polypeptide in Type 2 Diabetes Mellitus. J. Diabetes Res. 2016, 2016, 5639875.
- Bhowmick, D.C.; Kudaibergenova, Z.; Burnett, L.; Jeremic, A.M. Molecular Mechanisms of Amylin Turnover, Misfolding and Toxicity in the Pancreas. Molecules 2022, 27, 1021.
- Knight, J.D.; Miranker, A.D. Phospholipid catalysis of diabetic amyloid assembly. J. Mol. Biol. 2004, 341, 1175–1187.
- Cho, W.-J.; Trikha, S.; Jeremic, A.M. Cholesterol Regulates Assembly of Human Islet Amyloid Polypeptide on Model Membranes. J. Mol. Biol. 2009, 393, 765–775.
- Pivovarova, O.; Höhn, A.; Grune, T.; Pfeiffer, A.F.; Rudovich, N. Insulin-degrading enzyme: New therapeutic target for diabetes and Alzheimer’s disease? Ann. Med. 2016, 48, 614–624.
- Bennett, R.G.; Duckworth, W.C.; Hamel, F.G. Degradation of Amylin by Insulin-degrading Enzyme. J. Biol. Chem. 2000, 275, 36621–36625.
- Sousa, L.; Guarda, M.; Meneses, M.J.; Macedo, M.P.; Miranda, H.V. Insulin-degrading enzyme: An ally against metabolic and neurodegenerative diseases. J. Pathol. 2021, 255, 346–361.
- Bennett, R.G.; Hamel, F.G.; Duckworth, W.C. An Insulin-Degrading Enzyme Inhibitor Decreases Amylin Degradation, Increases Amylin-Induced Cytotoxicity, and Increases Amyloid Formation in Insulinoma Cell Cultures. Diabetes 2003, 52, 2315–2320.
- Hogan, M.F.; Zeman-Meier, D.; Zraika, S.; Templin, A.; Mellati, M.; Hull, R.L.; Leissring, M.A.; Kahn, S.E. Inhibition of Insulin-Degrading Enzyme Does Not Increase Islet Amyloid Deposition in Vitro. Endocrinology 2016, 157, 3462–3468.
- Maianti, J.P.; McFedries, A.; Foda, Z.H.; Kleiner, R.E.; Du, X.Q.; Leissring, M.A.; Tang, W.-J.; Charron, M.J.; Seeliger, M.A.; Saghatelian, A.; et al. Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones. Nature 2014, 511, 94–98.
- Tang, W.-J. Targeting Insulin-Degrading Enzyme to Treat Type 2 Diabetes Mellitus. Trends Endocrinol. Metab. 2016, 27, 24–34.
- Luca, S.; Yau, W.-M.; Leapman, R.; Tycko, R. Peptide Conformation and Supramolecular Organization in Amylin Fibrils: Constraints from Solid-State NMR. Biochemistry 2007, 46, 13505–13522.
- Wiltzius, J.J.; Sievers, S.A.; Sawaya, M.R.; Cascio, D.; Popov, D.; Riekel, C.; Eisenberg, D. Atomic structure of the cross-β spine of islet amyloid polypeptide (amylin). Protein Sci. 2008, 17, 1467–1474.
- Raleigh, D.; Zhang, X.; Hastoy, B.; Clark, A. The β-cell assassin: IAPP cytotoxicity. J. Mol. Endocrinol. 2017, 59, R121–R140.
- Hoffmann, K.Q.; McGovern, M.; Chiu, C.-C.; De Pablo, J.J. Secondary Structure of Rat and Human Amylin across Force Fields. PLoS ONE 2015, 10, e0134091.
- Wineman-Fisher, V.; Atsmon-Raz, Y.; Miller, Y. Orientations of Residues along the β-Arch of Self-Assembled Amylin Fibril-Like Structures Lead to Polymorphism. Biomacromolecules 2015, 16, 156–165.
- Matveyenko, A.V.; Butler, P.C. β-Cell Deficit Due to Increased Apoptosis in the Human Islet Amyloid Polypeptide Transgenic (HIP) Rat Recapitulates the Metabolic Defects Present in Type 2 Diabetes. Diabetes 2006, 55, 2106–2114.
- Krotee, P.; Rodriguez, J.A.; Sawaya, M.R.; Cascio, D.; Reyes, F.E.; Shi, D.; Hattne, J.; Nannenga, B.; Oskarsson, M.E.; Philipp, S.; et al. Atomic structures of fibrillar segments of hIAPP suggest tightly mated β-sheets are important for cytotoxicity. eLife 2017, 6, e19273.
- Zeman-Meier, D.; Entrup, L.; Templin, A.; Hogan, M.F.; Mellati, M.; Zraika, S.; Hull, R.L.; Kahn, S.E. The S20G substitution in hIAPP is more amyloidogenic and cytotoxic than wild-type hIAPP in mouse islets. Diabetologia 2016, 59, 2166–2171.
- Ridgway, Z.; Zhang, X.; Wong, A.G.; Abedini, A.; Schmidt, A.M.; Raleigh, D.P. Analysis of the Role of the Conserved Disulfide in Amyloid Formation by Human Islet Amyloid Polypeptide in Homogeneous and Heterogeneous Environments. Biochemistry 2018, 57, 3065–3074.
- Altamirano-Bustamante, M.M.; Altamirano-Bustamante, N.F.; Larralde-Laborde, M.; Lara-Martínez, R.; Leyva-García, E.; Garrido-Magaña, E.; Rojas, G.; Jiménez-García, L.F.; Monsalve, M.C.R.; Altamirano, P.; et al. Unpacking the aggregation-oligomerization-fibrillization process of naturally-occurring hIAPP amyloid oligomers isolated directly from sera of children with obesity or diabetes mellitus. Sci. Rep. 2019, 9, 18465.
- Burillo, J.; Fernández-Rhodes, M.; Piquero, M.; López-Alvarado, P.; Menéndez, J.; Jiménez, B.; González-Blanco, C.; Marqués, P.; Guillén, C.; Benito, M. Human amylin aggregates release within exosomes as a protective mechanism in pancreatic β cells: Pancreatic β-hippocampal cell communication. Biochim. Biophys. Acta 2021, 1868, 118971.
- Hiddinga, H.J.; Eberhardt, N.L. Intracellular Amyloidogenesis by Human Islet Amyloid Polypeptide Induces Apoptosis in COS-1 Cells. Am. J. Pathol. 1999, 154, 1077–1088.
- Bag, N.; Ali, A.; Chauhan, V.S.; Wohland, T.; Mishra, A. Membrane destabilization by monomeric hIAPP observed by imaging fluorescence correlation spectroscopy. Chem. Commun. 2013, 49, 9155–9157.
- Pilkington, E.; Gurzov, E.; Kakinen, A.; A Litwak, S.; Stanley, W.J.W.; Davis, T.P.T.; Ke, P.C. Pancreatic β-Cell Membrane Fluidity and Toxicity Induced by Human Islet Amyloid Polypeptide Species. Sci. Rep. 2016, 6, 21274.
- Engel, M.F.M.; Khemtémourian, L.; Kleijer, C.C.; Meeldijk, H.J.D.; Jacobs, J.; Verkleij, A.J.; de Kruijff, B.; Killian, J.A.; Höppener, J.W.M. Membrane damage by human islet amyloid polypeptide through fibril growth at the membrane. Proc. Natl. Acad. Sci. USA 2008, 105, 6033–6038.




