Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Deena Fayyad | -- | 2090 | 2022-04-27 04:06:28 | | | |
| 2 | Peter Tang | -5 word(s) | 2085 | 2022-04-27 04:43:07 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Fayyad, D.; Kelts, J.; Nielson, T.; , .; Xhabija, B. Viral Transmissibility of SARS-CoV-2. Encyclopedia. Available online: https://encyclopedia.pub/entry/22338 (accessed on 07 February 2026).
Fayyad D, Kelts J, Nielson T, , Xhabija B. Viral Transmissibility of SARS-CoV-2. Encyclopedia. Available at: https://encyclopedia.pub/entry/22338. Accessed February 07, 2026.
Fayyad, Deena, Jessica Kelts, Tristan Nielson, , Besa Xhabija. "Viral Transmissibility of SARS-CoV-2" Encyclopedia, https://encyclopedia.pub/entry/22338 (accessed February 07, 2026).
Fayyad, D., Kelts, J., Nielson, T., , ., & Xhabija, B. (2022, April 27). Viral Transmissibility of SARS-CoV-2. In Encyclopedia. https://encyclopedia.pub/entry/22338
Fayyad, Deena, et al. "Viral Transmissibility of SARS-CoV-2." Encyclopedia. Web. 27 April, 2022.
Copy Citation
The emergence of coronavirus disease 2019 (COVID-19), caused by the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to a global health calamity unprecedented in the modern world. The disease spread worldwide, and to date, there have been over 230 million confirmed cases of COVID-19, including approximately 4.7 million deaths. Mutant variants of the virus have raised concerns about additional pandemic waves and threaten to reverse our progress thus far to limit the spread of the virus.
COVID-19
SARS-CoV-2
viral transmissibility
1. Introduction
The emergence of the coronavirus disease 2019 (COVID-19), caused by the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to a global health calamity that is unprecedented in the modern world. Part of the Genus ‘Coronaviruses’ belonging to the family ‘Coronaviridae’, SARS-CoV-2 is the third virus of its kind responsible for an outbreak (albeit with greater global impact), succeeding SARS-CoV in 2003 and MERS-CoV (the Middle East Respiratory Syndrome Coronavirus) in 2012–2015 and 2020 [1]. COVID-19 emerged as novel pneumonia in Wuhan, Hubei Province, China, in December 2019 and is believed to have originated via zoonotic transmission. The disease quickly spread worldwide, prompting the World Health Organization (WHO) to declare the outbreak a pandemic on 11 March 2020. To date, there have been over 230 million confirmed cases of COVID-19, including approximately 4.7 million deaths [2]. The pandemic has also disrupted the global economic landscape due to widespread lockdowns, leading to loss of income for individuals and precipitating staggering negative trends in global stock markets [3].
2. Viral Transmissibility
The primary mode of transmission of SARS-CoV-2 is human-to-human via respiratory droplets produced during exhalation, as shown in Figure 1 [4][5][6]. However, there is also evidence that suggests the virus is transmissible via viral aerosol particles produced by individuals with COVID-19 disease [7][8][9]. In order to assist in the development of prevention guidelines, a group from the Nebraska Medical Center further investigated the transmission routes of the virus. Their studies reveal that viral RNA is present in nearly two-thirds of the air samples from rooms housing COVID-19 patients [9]. However, there is insufficient evidence at this time that would suggest that these particles indicate a viable virus that could be transmitted [9]. While the transmissibility of the virus via inanimate surfaces is generally low [10][11], aerosol to surface studies reveal that SARS-CoV-2 is more stable on plastic and stainless steel surfaces, showing a presence for up to several days, Figure 1 [12].
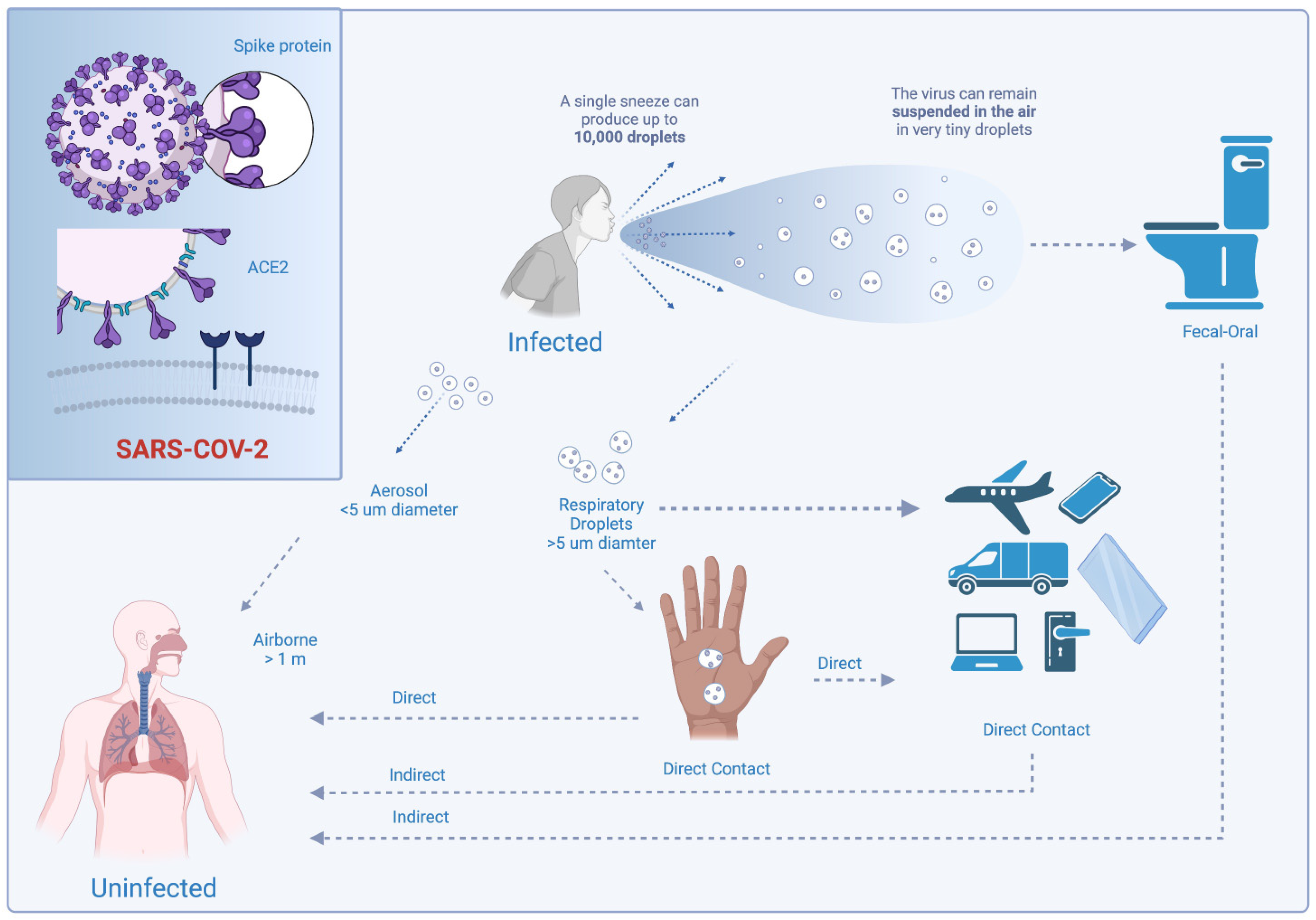
Figure 1. Modes of transmission of Sars-Cov-2. Created with BioRender.com.
An additional potential transmission route is fecal-oral transmission, Figure 1 [13][14][15][16]. Clinical characterization data from pediatric patients in China has suggested that rectal swab testing is more efficacious than nasopharyngeal testing. Several patients had tested negative for the nasopharyngeal swabs while positive for the rectal swabs [14][15][17]. However, the number of SARS-CoV-2 particles and the length of exposure necessary to cause infection are unknown [7].
Yet, the epidemiological and clinical findings indicate that the transmissibility of COVID-19 depends on the viral load at the time of the onset of the disease. Researchers have observed a short serial interval (time between symptom onset of the index case and secondary case) of 4–5 days for COVID-19 and a decreased risk for secondary infection over time. This suggests that there is a high risk of secondary transmission right before or at the onset of symptoms [4][18][19]. The recent data from South Korea reveals that the mean serial interval in the Delta variant is one day longer than when compared to the wild-type [20][21]. This trend is consistent with the viral shedding duration, as viral load peaks around symptom onset and decreases thereafter [4][18][22][23][24]. However, in more severe diseases, viral load was found to have peaked later in the second week of illness, suggesting a potentially longer period of infectiousness in cases of severe disease [25].
Recent discovery-based control design studies reveal that mutations in the viral nucleocapsid protein bring to light an association with the hospitalization rate [26]. Some single nucleotide variants increased the hospitalization rate, whereas a handful decreased it. Specifically, nonsynonymous variants R203K, R203S, and G204R occur in the linker region between the N-terminal RNA binding domain and the C-terminal dimerization domain, however this region has yet to be resolved [26][27]. It is hypothesized that the mutations in the linker region may potentially affect RNA binding interactions [26]. In addition, nucleocapsid mutations in the endoRNAse are also found to play an important role in the hospitalization status of the patients.
Several studies have examined the relationship between viral load and secondary transmission among symptomatic versus asymptomatic COVID-19 patients, although these findings have been inconsistent and contradictory. For instance, one study found comparable viral load in nasal and throat swabs at symptom onset among symptomatic and asymptomatic patients [24], while another found higher viral load in nasopharyngeal swabs in symptomatic patients [18]. Notably, a study by Hasanoglu et al. identified significantly higher viral load in asymptomatic patients in six types of specimens (nasopharyngeal/oropharyngeal, oral cavity, saliva, rectal, urine, and blood), suggesting higher infectiousness for asymptomatic patients than previously thought [28]. These findings highlight an important gap in the literature regarding the viral load and transmission risk of symptomatic versus asymptomatic COVID-19 patients that warrant further investigation.
Similarly, there are a limited number of studies in the literature that assess the relationship between SARS-CoV-2 viral load and disease severity. As was previously stated, disease severity may be associated with a higher risk of secondary transmission; however, this hypothesis must be examined further. Nonetheless, several studies have identified an association between disease severity and SARS-CoV-2 RNA shedding [29][30][31], while others report no such correlation [8][16]. For instance, the longer the duration of SARS-CoV-2 RNA shedding, the longer it takes to recover body temperature (when the fever was present at illness onset) compared to patients with early SARS-CoV-2 RNA clearance [31]. Additionally, patients admitted to the Intense Care Unit experienced longer viral shedding time than non-ICU patients [32]. A similar pattern is identified for the relative amounts of viral load over time, where higher viral load is associated with increased disease severity and mortality [33][34]. However, other studies have found that viral load in severe patients is lower [28] than in mild cases, encouraging further research in this area.
One possible reason for these discrepancies in the literature relating to viral load and transmissibility may be due to the wide range of reported viral shedding times for SARS-CoV-2. For instance, the average reported duration of viral shedding from the onset of the illness ranges from 11 days [29] to 17 days [31] to 31 days [35]. The median duration of viral shedding also differs in the type of specimen collected. Major shedding routes for SARS-CoV-2, such as the nasopharyngeal, sputum, and stools [30], are expected to show a longer median duration of viral shedding than other collected specimens. Interestingly, sputum specimens exhibit longer viral shedding than nasopharyngeal specimens [30][36], almost twice as long according to some accounts [36]. Hindson et al., 2020 determined that the digestive system may exhibit longer viral shedding than the respiratory tract [14]. These findings are supported by the fact that patients with COVID-19 can simultaneously test positive for SARS-CoV-2 RNA on some samples but not on others. For instance, patients simultaneously tested positive for COVID-19 on rectal swabs but tested negative on nasopharyngeal swabs within the same testing period [13][14]. COVID-19 patients may also test negative for SARS-CoV-2 on samples that later test positive [14][37], further limiting the generalizability of viral shedding times in the literature.
There are other factors that have been associated with the transmission. One of them is individuals’ behaviors toward public health measures, such as physically distancing, masking, or staying home while they are sick. For example, those who had lower concerns about spreading the virus reflected the least uptake of public health measures [38]. Transmission dynamics were also affected among close contacts. Specifically, the transmission potential was at its peak in the first two days and after three days of the onset of the symptoms. When individuals were exposed to mild and moderate COVID-19 patients, they were exposed to a higher risk of COVID-19 when compared to asymptomatic carriers, and it worsens when exposed to patients with moderate cases of COVID-19 [39]. Moreover, environmental conditions play an important two-fold role; the initial viral load, and the immune response. Generally, higher temperatures and humidity have been associated with a lower fatality rate [40]. Another factor that has played an important role in Sars-CoV-2 transmission is long diagnostic delays (LDDs). A study in Japan concluded that the portion of long diagnostic delays with unknown exposure was correlated with a significant increase in the virus spread [41].
Moreover, the range of reported viral shedding times of SARS-CoV-2 may also be explained by independent risk factors such as age and gender. In some reports, old age—a major risk factor for COVID-19 severity—is associated with prolonged viral shedding duration [31][36]. Contrary to Xu K et al., 2020 and Wang K et al., 2020, another study finds no significant difference in viral shedding time between patients less than 65 years of age and those aged 65 years and older [35]. Similarly, some studies find viral shedding duration in males to be higher [31] or equal to females [35]. Concomitant hypertension has also been identified as a potential risk factor for prolonged viral shedding [31].
Conversely, the differences in the duration of viral shedding may be due to features that vary on a case-by-case basis, such as a longer time from the onset of symptoms to hospitalization or treatment, which increases the risk for prolonged viral shedding [29][31][42]. Other factors that are associated with an increased duration of viral shedding include cough [30], fever, and hydrocortisone use [29], specifically high-dose corticosteroids [43]. The presence of diseases other than COVID-19, such as diabetes mellitus and chronic lung disease, is associated with viral RNA detection [18]. Overall, these studies highlight the need for additional research on the factors affecting viral shedding duration and the detection of SARS-CoV-2.
As is the case with other coronaviruses, SARS-CoV-2 relies on the spike (S) glycoprotein to successfully bind to and enter host cells. Components of the SARS-CoV-2 S protein include subunit S1, subunit S2, the transmembrane anchor, and the intracellular trail [44][45][46]. However, while SARS-CoV-2 related coronaviruses today contain a monobasic cleavage site between S1 and S2, SARS-CoV-2 garners a multibasic cleavage site, Figure 2 [47][48][49], believed to be the result of a recombination event [16][35][47]. This specialized motif enables SARS-CoV-2 to exploit a greater variety of widespread cellular proteases in the body, thereby allowing the virus to have a more rapid spread [44][46]. Additionally, newly synthesized virions can bypass the requirement of binding to host cell receptors and still continue to infect cells as they can be secreted in a preactivated state [46].
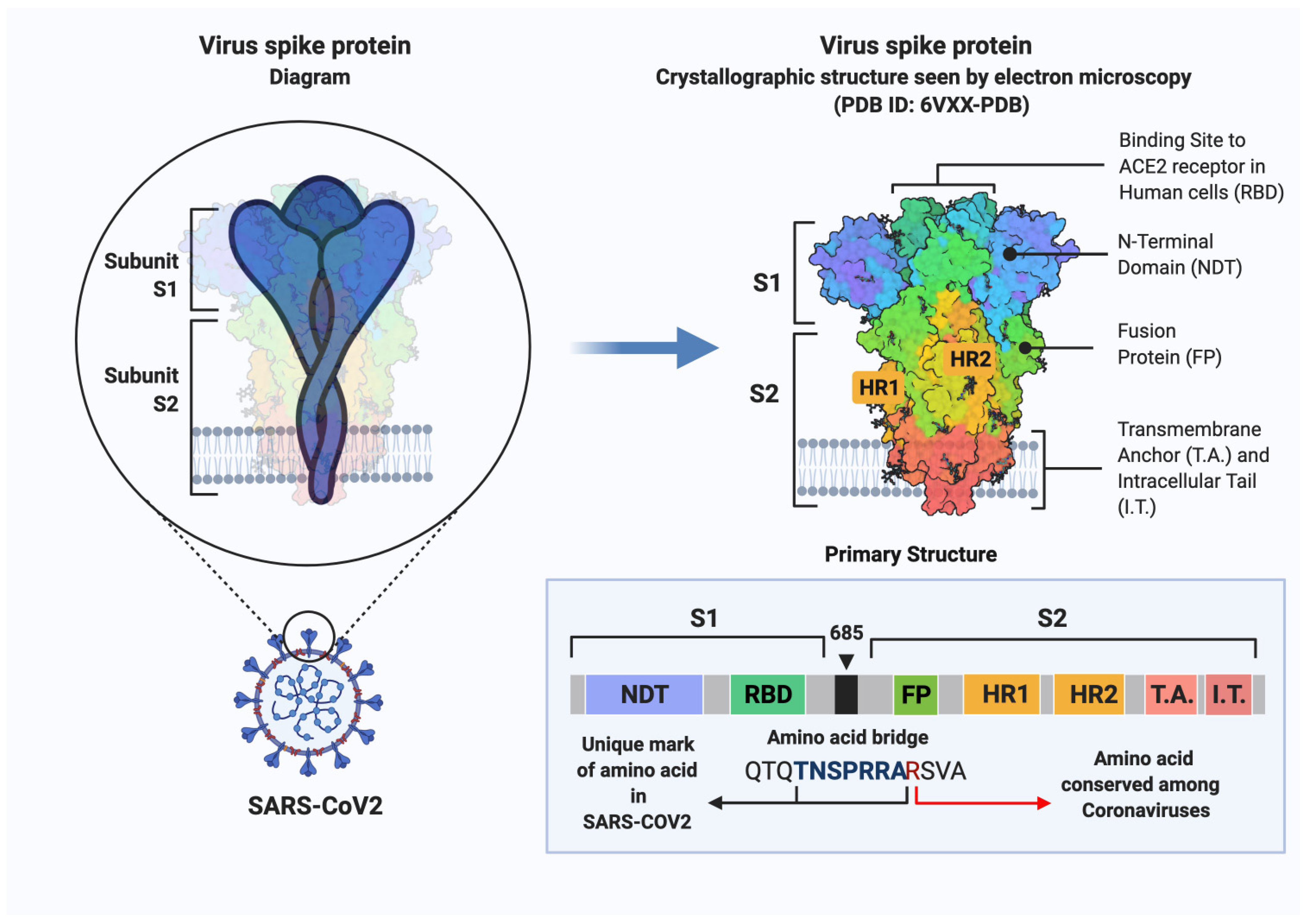
Figure 2. Schematic representation of SARS-CoV-2 spike protein primary structure. Domains are depicted with different colors. NTE, N-terminal domain; RBD, receptor-binding domain; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain; IT, intracellular tail. Created with BioRender.com.
Moreover, the SARS-CoV-2 S protein can trigger cellular entry independent of proteases but dependent on receptor binding, leading to augmented viral spread [46]. However, when the S1/S2 site is damaged, it can severely inhibit the S protein cleavage and proteolytic processing [47]. Moreover, structural studies suggest that the addition of basic residues to the S1/S2 site of SARS-CoV-2 exhibits an increased viral spread via cell–cell fusion but no change to virus–cell fusion [47]. In sum, the structural studies stress the importance of recognizing which viral mutants lead to inhibition or augmentation of viral spread to develop effective therapeutic strategies against SARS-CoV-2 and its variants.
For viral spread to occur, the S protein of SARS-CoV-2 must first be activated. This process starts when the receptor-binding domain (RBD) of the S1 subunit binds to the host cell surface receptor angiotensin-converting enzyme-2 (ACE2) [44][46][47][50] via its peptidase domain [51][52]. Compared to SARS-CoV, the receptor binding capacity of SARS-CoV-2 to ACE2 is at least 10-fold higher [44][49]. This high affinity is partially due to the specialized RBD of SARS-CoV-2, which contains a residue motif at 482–485 (Gly-Val-Glu-Gly) that allows for better contact with ACE2 and two key residues (Gln493 and Leu455) that stabilize ACE2 binding [46][53]. Interestingly, the data reveals that this high-affinity results in increased virulence of SARS-CoV-2 [4][48][49]. Once SARS-CoV-2 and ACE2 bind together, it alters the conformation of the S protein, exposing a cleavage site on the S2 subunit, which the host cell proteases will act upon in the next step [46][54]. Then, the host cell proteases, such as transmembrane protease serine S1 member 2 (TMPRSS2), carry out proteolysis of the S protein at the cleavage site between the S1/S2 boundary, resulting in a new S2 site (S2′) [46][55][56]. Following the S2′ cleavage and the release of the S2 subunit, the activation of the protein concludes, allowing for the subsequent initiation of the endocytosis of the virus through the fusion of the viral and cell membranes [46][54][55][56].
References
- Balasubramanian Ganesh; Thangarasu Rajakumar; Mathiyazhakan Malathi; Natesan Manikandan; Jaganathasamy Nagaraj; Aridoss Santhakumar; Arumugam Elangovan; Yashpal Singh Malik; Epidemiology and pathobiology of SARS-CoV-2 (COVID-19) in comparison with SARS, MERS: An updated overview of current knowledge and future perspectives. Clinical Epidemiology and Global Health 2021, 10, 100694-100694, 10.1016/j.cegh.2020.100694.
- WHO Coronavirus (COVID-19) Dashboard . World Health Organization. Retrieved 2022-4-26
- Maria Nicola; Zaid Alsafi; Catrin Sohrabi; Ahmed Kerwan; Ahmed Al-Jabir; Christos Iosifidis; Maliha Agha; Riaz Agha; The socio-economic implications of the coronavirus pandemic (COVID-19): A review. International Journal of Surgery 2020, 78, 185-193, 10.1016/j.ijsu.2020.04.018.
- Jasper Fuk-Woo Chan; Shuofeng Yuan; Kin-Hang Kok; Kelvin Kai-Wang To; Hin Chu; Jin Yang; Fanfan Xing; Jieling Liu; Cyril Chik-Yan Yip; Rosana Wing-Shan Poon; et al.Hoi-Wah TsoiSimon Kam-Fai LoKwok-Hung ChanVincent Kwok-Man PoonWan-Mui ChanJonathan Daniel IpJian-Piao CaiVincent Chi-Chung ChengHonglin ChenChristopher Kim-Ming HuiKwok-Yung Yuen A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. The Lancet 2020, 395, 514-523, 10.1016/s0140-6736(20)30154-9.
- Rutu Karia; Ishita Gupta; Harshwardhan Khandait; Ashima Yadav; Anmol Yadav; COVID-19 and its Modes of Transmission. SN Comprehensive Clinical Medicine 2020, 2, 1798-1801, 10.1007/s42399-020-00498-4.
- Andrew G. Harrison; Tao Lin; Penghua Wang; Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends in Immunology 2020, 41, 1100-1115, 10.1016/j.it.2020.10.004.
- Is the coronavirus airborne? Experts can’t agree . Nature. Retrieved 2022-4-26
- Grace Lui; Lowell Ling; Christopher Lai; Eugene Yk Tso; Kitty Sc Fung; Veronica Chan; Tracy Hy Ho; Fion Luk; Zigui Chen; Joyce Kc Ng; et al.Kai-Ming ChowPeter Kc ChengRickjason Cw ChanDominic Nc TsangCharles D GomersallDavid Sc HuiPaul Ks Chan Viral dynamics of SARS-CoV-2 across a spectrum of disease severity in COVID-19. Journal of Infection 2020, 81, 318-356, 10.1016/j.jinf.2020.04.014.
- Joshua L. Santarpia; Danielle N. Rivera; Vicki L. Herrera; M. Jane Morwitzer; Hannah Creager; George W. Santarpia; Kevin K. Crown; David M. Brett-Major; Elizabeth R. Schnaubelt; M. Jana Broadhurst; et al.James V. LawlerSt. Patrick ReidJohn J. Lowe Aerosol and surface contamination of SARS-CoV-2 observed in quarantine and isolation care. Scientific Reports 2020, 10, 1-8, 10.1038/s41598-020-69286-3.
- Emanuel Goldman; Exaggerated risk of transmission of COVID-19 by fomites. The Lancet Infectious Diseases 2020, 20, 892-893, 10.1016/s1473-3099(20)30561-2.
- Mario U Mondelli; Marta Colaneri; Elena M Seminari; Fausto Baldanti; Raffaele Bruno; Low risk of SARS-CoV-2 transmission by fomites in real-life conditions. The Lancet Infectious Diseases 2020, 21, e112, 10.1016/s1473-3099(20)30678-2.
- Neeltje van Doremalen; Trenton Bushmaker; Dylan H. Morris; Myndi G. Holbrook; Amandine Gamble; Brandi N. Williamson; Azaibi Tamin; Jennifer L. Harcourt; Natalie J. Thornburg; Susan I. Gerber; et al.James O. Lloyd-SmithEmmie de WitVincent J. Munster Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. New England Journal of Medicine 2020, 382, 1564-1567, 10.1056/nejmc2004973.
- Yifei Chen; Liangjun Chen; Qiaoling Deng; Guqin Zhang; Kaisong Wu; Lan Ni M.D.; Yibin Yang; Bing Liu; Wei Wang; Chaojie Wei; et al.Jiong YangGuangming YeZhenshun Cheng The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. Journal of Medical Virology 2020, 92, 833-840, 10.1002/jmv.25825.
- Jordan Hindson; COVID-19: faecal-oral transmission?. Nature Reviews Gastroenterology & Hepatology 2020, 17, 259-259, 10.1038/s41575-020-0295-7.
- Yongjian Wu; Cheng Guo; Lantian Tang; Zhongsi Hong; Jianhui Zhou; Xin Dong; Huan Yin; Qiang Xiao; Yanping Tang; Xiujuan Qu; et al.Liangjian KuangXiaomin FangNischay MishraJiahai LuHong ShanGuanmin JiangXi Huang Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. The Lancet Gastroenterology & Hepatology 2020, 5, 434-435, 10.1016/s2468-1253(20)30083-2.
- Xinchun Zheng; Jiehua Chen; Lisi Deng; Zhaoxiong Fang; Gongqi Chen; Di Ye; Jinyu Xia; Zhongsi Hong; Risk factors for the COVID‐19 severity and its correlation with viral shedding: A retrospective cohort study. Journal of Medical Virology 2020, 93, 952-961, 10.1002/jmv.26367.
- Yi Xu; Xufang Li; Bing Zhu; Huiying Liang; Chunxiao Fang; Yu Gong; Qiaozhi Guo; Xin Sun; Danyang Zhao; Jun Shen; et al.Huayan ZhangHongsheng LiuHuimin XiaJinling TangKang ZhangSitang Gong Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nature Medicine 2020, 26, 502-505, 10.1038/s41591-020-0817-4.
- Hitoshi Kawasuji; Yusuke Takegoshi; Makito Kaneda; Akitoshi Ueno; Yuki Miyajima; Koyomi Kawago; Yasutaka Fukui; Yoshihiro Yoshida; Miyuki Kimura; Hiroshi Yamada; et al.Ippei SakamakiHideki TaniYoshitomo MorinagaYoshihiro Yamamoto Transmissibility of COVID-19 depends on the viral load around onset in adult and symptomatic patients. PLoS ONE 2020, 15, e0243597, 10.1371/journal.pone.0243597.
- Hiroshi Nishiura; Natalie M. Linton; Andrei R. Akhmetzhanov; Serial interval of novel coronavirus (COVID-19) infections. International Journal of Infectious Diseases 2020, 93, 284-286, 10.1016/j.ijid.2020.02.060.
- Meng Zhang; Guangzhou Guangdong Provincial Center For Disease Control And Prevention; Jianpeng Xiao; Aiping Deng; Yingtao Zhang; Yali Zhuang; Ting Hu; Jiansen Li; Hongwei Tu; Bosheng Li; et al.Yan ZhouJun YuanLei LuoZimian LiangYouzhi HuangGuoqiang YeMingwei CaiGongli LiBo YangBin XuXiming HuangYazun CuiDongsheng RenYanping ZhangMin KangYan LiBeijing Chinese Center For Disease Control And Prevention Transmission Dynamics of an Outbreak of the COVID-19 Delta Variant B.1.617.2—Guangdong Province, China, May-June 2021. China CDC Weekly 2020, 3, 584-586, 10.46234/ccdcw2021.148.
- Sukhyun Ryu; Dasom Kim; Jun-Sik Lim; Sheikh Taslim Ali; Benjamin J. Cowling; Serial Interval and Transmission Dynamics during SARS-CoV-2 Delta Variant Predominance, South Korea. Emerging Infectious Diseases 2022, 28, 407-410, 10.3201/eid2802.211774.
- Xi He; Eric H. Y. Lau; Peng Wu; Xilong Deng; Jian Wang; Xinxin Hao; Yiu Chung Lau; Jessica Y. Wong; Yujuan Guan; Xinghua Tan; et al.Xiaoneng MoYanqing ChenBaolin LiaoWeilie ChenFengyu HuQing ZhangMingqiu ZhongYanrong WuLingZhai ZhaoFuchun ZhangBenjamin J. CowlingFang LiGabriel M. Leung Author Correction: Temporal dynamics in viral shedding and transmissibility of COVID-19. Nature Medicine 2020, 26, 1491-1493, 10.1038/s41591-020-1016-z.
- Kelvin Kai-Wang To; Owen Tak-Yin Tsang; Wai-Shing Leung; Anthony Raymond Tam; Tak-Chiu Wu; David Christopher Lung; Cyril Chik-Yan Yip; Jian-Piao Cai; Jacky Man-Chun Chan; Thomas Shiu-Hong Chik; et al.Daphne Pui-Ling LauChris Yau-Chung ChoiLin-Lei ChenWan-Mui ChanKwok-Hung ChanJonathan Daniel IpAnthony Chin-Ki NgRosana Wing-Shan PoonCui-Ting LuoVincent Chi-Chung ChengJasper Fuk-Woo ChanIvan Fan-Ngai HungZhiwei ChenHonglin ChenKwok-Yung Yuen Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. The Lancet Infectious Diseases 2020, 20, 565-574, 10.1016/s1473-3099(20)30196-1.
- Lirong Zou; Feng Ruan; Mingxing Huang; Lijun Liang; Huitao Huang; Zhongsi Hong; Jianxiang Yu; Min Kang; Yingchao Song; Jinyu Xia; et al.Qianfang GuoTie SongJianfeng HeHui-Ling YenMalik PeirisJie Wu SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. New England Journal of Medicine 2020, 382, 1177-1179, 10.1056/nejmc2001737.
- Yuan Liu; Zhi Ning; Yu Chen; Ming Guo; Yingle Liu; Nirmal Kumar Gali; Li Sun; Yusen Duan; Jing Cai; Dane Westerdahl; et al.Xinjin LiuKe XuKin-Fai HoHaidong KanQingyan FuKe Lan Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature 2020, 582, 557-560, 10.1038/s41586-020-2271-3.
- Lue Ping Zhao; Pavitra Roychoudhury; Peter Gilbert; Joshua Schiffer; Terry P. Lybrand; Thomas H. Payne; April Randhawa; Sara Thiebaud; Margaret Mills; Alex Greninger; et al.Chul-Woo PyoRuihan WangRenyu LiAlexander ThomasBrandon NorrisWyatt C. NelsonKeith R. JeromeDaniel E. Geraghty Mutations in viral nucleocapsid protein and endoRNase are discovered to associate with COVID19 hospitalization risk. Scientific Reports 2022, 12, 1-11, 10.1038/s41598-021-04376-4.
- Weihong Zeng; Guangfeng Liu; Huan Ma; Dan Zhao; Yunru Yang; Muziying Liu; Ahmed Mohammed; Changcheng Zhao; Yun Yang; Jiajia Xie; et al.Chengchao DingXiaoling MaJianping WengYong GaoHongliang HeTengchuan Jin Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochemical and Biophysical Research Communications 2020, 527, 618-623, 10.1016/j.bbrc.2020.04.136.
- Imran Hasanoglu; Gulay Korukluoglu; Dilek Asilturk; Yasemin Cosgun; Ayse Kaya Kalem; Ayşe Basak Altas; Bircan Kayaaslan; Fatma Eser; Esra Akkan Kuzucu; Rahmet Guner; et al. Higher viral loads in asymptomatic COVID-19 patients might be the invisible part of the iceberg. Infection 2020, 49, 117-126, 10.1007/s15010-020-01548-8.
- Tong‐Zeng Li; Zhen‐Huan Cao; Yu Chen; Miao‐Tian Cai; Long‐Yu Zhang; Hui Xu; Jia‐Ying Zhang; Chun‐Hua Ma; Yang Liu; Li‐Juan Gao; et al.Zhong‐Hui DuanDan‐Lei MouLian‐Chun Liang Duration of SARS-CoV-2 RNA shedding and factors associated with prolonged viral shedding in patients with COVID-19. Journal of Medical Virology 2020, 93, 506-512, 10.1002/jmv.26280.
- Wenting Tan; Yanqiu Lu; Juan Zhang; Jing Wang; Yunjie Dan; Zhaoxia Tan; Xiaoqing He; Chunfang Qian; Qiangzhong Sun; Qingli Hu; et al.Honglan LiuSikuan YeXiaomei XiangYi ZhouWei ZhangYanzhi GuoXiu-Hua WangWeiwei HeXing WanFengming SunQuanfang WeiCong ChenGuangqiang PanJie XiaQing MaoYaokai ChenGuohong Deng Viral Kinetics and Antibody Responses in Patients with COVID-19. null 2020, null, null, 10.1101/2020.03.24.20042382.
- Kaijin Xu; Yanfei Chen; Jing Yuan; Ping Yi; Cheng Ding; Wenrui Wu; Yongtao Li; Qin Ni; Rongrong Zou; Xiaohe Li; et al.Min XuYing ZhangHong ZhaoXuan ZhangLiang YuJunwei SuGuanjing LangJun LiuXiaoxin WuYongzheng GuoJingjing TaoDing ShiQing CaoBing RuanLei LiuZhaoqin WangYan XuYingxia LiuJifang ShengLanjuan Li Factors Associated With Prolonged Viral RNA Shedding in Patients with Coronavirus Disease 2019 (COVID-19). Clinical Infectious Diseases 2020, 71, 799-806, 10.1093/cid/ciaa351.
- Zhixiong Fang; Yi Zhang; Changfa Hang; Jingwen Ai; Shaojie Li; Wenhong Zhang; Comparisons of viral shedding time of SARS-CoV-2 of different samples in ICU and non-ICU patients. Journal of Infection 2020, 81, 147-178, 10.1016/j.jinf.2020.03.013.
- Jesse Fajnzylber; The Massachusetts Consortium for Pathogen Readiness; James Regan; Kendyll Coxen; Heather Corry; Colline Wong; Alexandra Rosenthal; Daniel Worrall; Francoise Giguel; Alicja Piechocka-Trocha; et al.Caroline AtyeoStephanie FischingerAndrew ChanKeith T. FlahertyKathryn HallMichael DouganEdward T. RyanElizabeth GillespieRida ChishtiYijia LiNikolaus JilgDusan HanidziarRebecca M. BaronLindsey BadenAthe M. TsibrisKatrina A. ArmstrongDaniel R. KuritzkesGalit AlterBruce D. WalkerXu YuJonathan Z. Li SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nature Communications 2020, 11, 1-9, 10.1038/s41467-020-19057-5.
- Elisabet Pujadas; Fayzan Chaudhry; Russell McBride; Felix Richter; Shan Zhao; Ania Wajnberg; Girish Nadkarni; Benjamin S Glicksberg; Jane Houldsworth; Carlos Cordon-Cardo; et al. SARS-CoV-2 viral load predicts COVID-19 mortality. The Lancet Respiratory Medicine 2020, 8, e70-e70, 10.1016/s2213-2600(20)30354-4.
- Bo Zhou; Jianqing She; Yadan Wang; Xiancang Ma; Duration of Viral Shedding of Discharged Patients With Severe COVID-19. Clinical Infectious Diseases 2020, 71, 2240-2242, 10.1093/cid/ciaa451.
- Kun Wang; Xin Zhang; Jiaxing Sun; Jia Ye; Feilong Wang; Jing Hua; Huayu Zhang; Ting Shi; Qiang Li; Xiaodong Wu; et al. Differences of Severe Acute Respiratory Syndrome Coronavirus 2 Shedding Duration in Sputum and Nasopharyngeal Swab Specimens Among Adult Inpatients With Coronavirus Disease 2019. Chest 2020, 158, 1876-1884, 10.1016/j.chest.2020.06.015.
- Lan Lan; Dan Xu; Guangming Ye; Chen Xia; Shaokang Wang; Yirong Li; Haibo Xu; Positive RT-PCR Test Results in Patients Recovered From COVID-19. JAMA 2020, 323, 1502, 10.1001/jama.2020.2783.
- Raynell Lang; Jamie L. Benham; Omid Atabati; Aidan Hollis; Trevor Tombe; Blake Shaffer; Katharina Kovacs Burns; Gail MacKean; Tova Léveillé; Brandi McCormack; et al.Hasan SheikhMadison M. FullertonTheresa TangJean-Christophe BoucherCora ConstantinescuMehdi MouraliBraden J. MannsDeborah A. MarshallJia HuRobert J. Oxoby Attitudes, behaviours and barriers to public health measures for COVID-19: a survey to inform public health messaging. BMC Public Health 2021, 21, 1-15, 10.1186/s12889-021-10790-0.
- Yang Ge; Leonardo Martinez; Shengzhi Sun; Zhiping Chen; Feng Zhang; Fangyu Li; Wanwan Sun; Enfu Chen; Jinren Pan; Changwei Li; et al.Jimin SunAndreas HandelFeng LingYe Shen COVID-19 Transmission Dynamics Among Close Contacts of Index Patients With COVID-19. JAMA Internal Medicine 2021, 181, 1343, 10.1001/jamainternmed.2021.4686.
- Jingbo Liang; Hsiang-Yu Yuan; Assessing the impact of temperature and humidity exposures during early infection stages on case-fatality of COVID-19: A modelling study in Europe. Environmental Research 2022, 211, 112931, 10.1016/j.envres.2022.112931.
- Tsuyoshi Ogata; Hideo Tanaka; Long Diagnostic Delay with Unknown Transmission Route Inversely Correlates with the Subsequent Doubling Time of Coronavirus Disease 2019 in Japan, February–March 2020. International Journal of Environmental Research and Public Health 2021, 18, 3377, 10.3390/ijerph18073377.
- Jing Han; Li-Xia Shi; Yi Xie; Yong-Jin Zhang; Shu-Ping Huang; Jian-Guo Li; He-Rong Wang; Shi-Feng Shao; Analysis of factors affecting the prognosis of COVID-19 patients and viral shedding duration. Epidemiology and Infection 2020, 148, e125, 10.1017/s0950268820001399.
- Sijia Li; Zhigang Hu; Xinyu Song; High-dose but Not Low-dose Corticosteroids Potentially Delay Viral Shedding of Patients With COVID-19. Clinical Infectious Diseases 2020, 72, 1297-1298, 10.1093/cid/ciaa829.
- Koa Hosoki; Abhijit Chakraborty; Sanjiv Sur; Molecular mechanisms and epidemiology of COVID-19 from an allergist’s perspective. Journal of Allergy and Clinical Immunology 2020, 146, 285-299, 10.1016/j.jaci.2020.05.033.
- Paul S. Masters; The Molecular Biology of Coronaviruses. Advances in Applied Microbiology 2006, 66, 193-292, 10.1016/s0065-3527(06)66005-3.
- Tahir S Pillay; Gene of the month: the 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. Journal of Clinical Pathology 2020, 73, 366-369, 10.1136/jclinpath-2020-206658.
- Markus Hoffmann; Hannah Kleine-Weber; Stefan Pöhlmann; A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Molecular Cell 2020, 78, 779-784.e5, 10.1016/j.molcel.2020.04.022.
- Alexandra C. Walls; Young-Jun Park; M. Alejandra Tortorici; Abigail Wall; Andrew T. McGuire; David Veesler; Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 183, 1735-1735, 10.1016/j.cell.2020.11.032.
- Daniel Wrapp; Nianshuang Wang; Kizzmekia S. Corbett; Jory A. Goldsmith; Ching-Lin Hsieh; Olubukola Abiona; Barney S. Graham; Jason S. McLellan; Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260-1263, 10.1126/science.abb2507.
- Qihui Wang; Yanfang Zhang; Lili Wu; Sheng Niu; Chunli Song; Zengyuan Zhang; Guangwen Lu; Chengpeng Qiao; Yu Hu; Kwok-Yung Yuen; et al.Qisheng WangHuan ZhouJinghua YanJianxun Qi Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894-904.e9, 10.1016/j.cell.2020.03.045.
- Fang Li; Wenhui Li; Michael Farzan; Stephen C. Harrison; Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science 2005, 309, 1864-1868, 10.1126/science.1116480.
- Wenhui Li; Chengsheng Zhang; Jianhua Sui; Jens H. Kuhn; Michael J Moore; Shiwen Luo; Swee-Kee Wong; I-Chueh Huang; Keming Xu; Natalya Vasilieva; et al.Akikazu MurakamiYaqing HeWayne A MarascoYi GuanHyeryun ChoeMichael Farzan Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. The EMBO Journal 2005, 24, 1634-1643, 10.1038/sj.emboj.7600640.
- Jian Shang; Gang Ye; Ke Shi; Yushun Wan; Chuming Luo; Hideki Aihara; Qibin Geng; Ashley Auerbach; Fang Li; Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221-224, 10.1038/s41586-020-2179-y.
- Markus Hoffmann; Heike Hofmann-Winkler; Stefan Pöhlmann; Priming Time: How Cellular Proteases Arm Coronavirus Spike Proteins. Activation of Viruses by Host Proteases 2018, null, 71-98, 10.1007/978-3-319-75474-1_4.
- Sandrine Belouzard; Victor C. Chu; Gary R. Whittaker; Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proceedings of the National Academy of Sciences 2009, 106, 5871-5876, 10.1073/pnas.0809524106.
- Jean Kaoru Millet; Gary R. Whittaker; Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proceedings of the National Academy of Sciences 2014, 111, 15214-15219, 10.1073/pnas.1407087111.
More
Information
Subjects:
Virology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
987
Entry Collection:
COVID-19
Revisions:
2 times
(View History)
Update Date:
27 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




