Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Emmanuel Jairaj Moses | -- | 1946 | 2022-04-21 05:59:52 | | | |
| 2 | Dean Liu | + 180 word(s) | 2126 | 2022-04-22 03:34:44 | | | | |
| 3 | Dean Liu | -7 word(s) | 2119 | 2022-04-22 03:40:15 | | | | |
| 4 | Dean Liu | Meta information modification | 2119 | 2022-04-22 03:40:54 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Moses, E.; , .; Rajasegaran, Y.; Heidenreich, O.; Mohd Yusoff, N.; Mot, Y. Mechanisms of microRNA Dysregulation in Cancer. Encyclopedia. Available online: https://encyclopedia.pub/entry/22065 (accessed on 07 February 2026).
Moses E, , Rajasegaran Y, Heidenreich O, Mohd Yusoff N, Mot Y. Mechanisms of microRNA Dysregulation in Cancer. Encyclopedia. Available at: https://encyclopedia.pub/entry/22065. Accessed February 07, 2026.
Moses, Emmanuel, , Yaashini Rajasegaran, Olaf Heidenreich, Narazah Mohd Yusoff, Yik Mot. "Mechanisms of microRNA Dysregulation in Cancer" Encyclopedia, https://encyclopedia.pub/entry/22065 (accessed February 07, 2026).
Moses, E., , ., Rajasegaran, Y., Heidenreich, O., Mohd Yusoff, N., & Mot, Y. (2022, April 21). Mechanisms of microRNA Dysregulation in Cancer. In Encyclopedia. https://encyclopedia.pub/entry/22065
Moses, Emmanuel, et al. "Mechanisms of microRNA Dysregulation in Cancer." Encyclopedia. Web. 21 April, 2022.
Copy Citation
Similar to genes, microRNA are also subjected to dysregulation, therefore various mechanisms need to be considered when studying miRNA dysregulation. Amongst them are the amplification of loci which leads to the overexpression of amplified genes, mutation leading to deregulated or loss of function, epigenetic control contributing to gene activation or suppression and the effects of trans activating elements on gene expressional control.
microRNA
miRNASponges
antagomir
miRNA mimic
1. Amplification of Loci
MicroRNA loci amplification is a common occurrence in various cancers (Figure 1). Amplification of the genomic region often causes overexpression of the miRNAs thus resulting in the depletion of its target mRNA [1]. This in turn leads to dysregulation in critical signaling pathways which contribute to cancer development and progression.
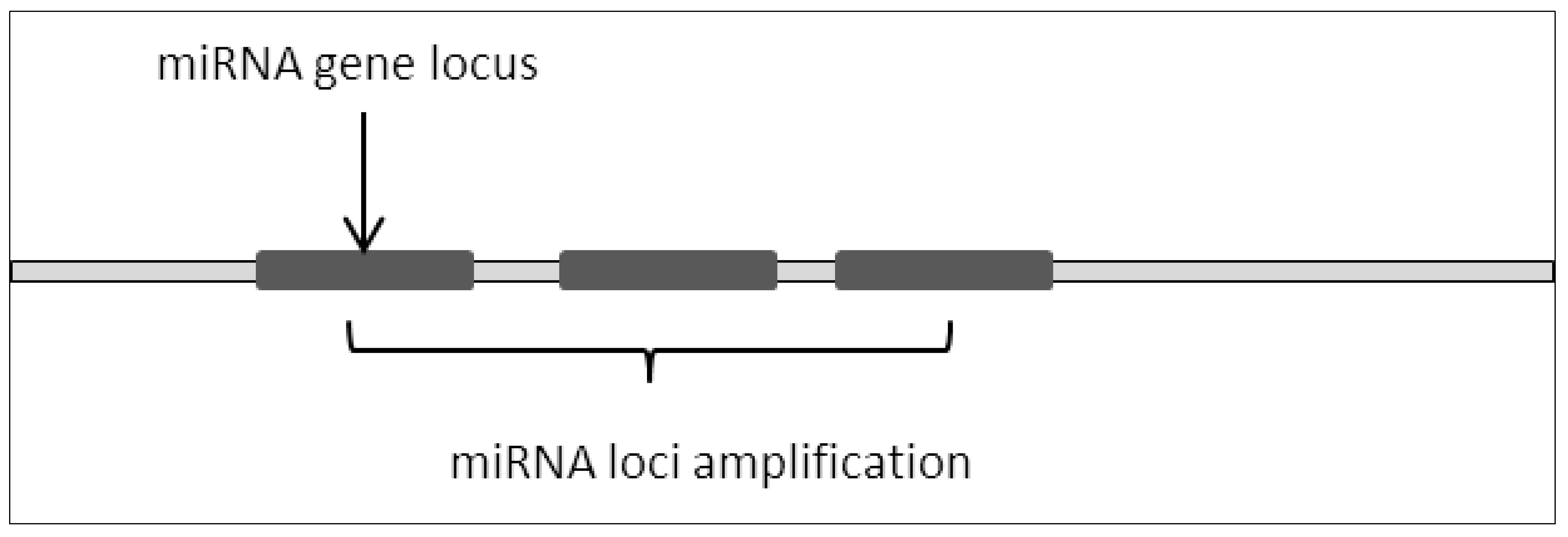
Figure 1. miRNA loci amplification resulted in repeating miRNA sequences thus increasing basal expression of miRNA.
This can be observed in embryonic brain tumors where the C19 miRNA cluster was amplified [2][3]. Interestingly, the same research also showed that the C19 cluster was fused to the TTYH1 gene, which further drives the expression of the amplified C19 cluster polycistronically. This causes extreme overexpression of DNM3TB which is an effector of the C19 miRNA cluster target, RBL2. It clearly shows that microRNA loci amplification leads to the dysregulation of target genes and their effectors.
Amplification also occurred in the miR-17-92 cluster where it was found that the miRNAs within the cluster were significantly overexpressed in human lung cancer [4]. MiR-17-92 cluster copy number was significantly higher when compared to the copy number of miR-106a-92 and miR 106b-25. Additionally, the same cluster was also found to be amplified in acute myeloid leukemia with MLL rearrangement where it was found that this cluster affects cellular differentiation and proliferation [5].
Furthermore, miR-23a locus was also found to be implicated in the same manner in gastric cancer. Research found that the miRNA was over-expressed and that the miRNA gene was amplified along with other miRNAs (miR-1274a, miR-196b, miR-4298, miR-181c, miR-181d, miR-23a, miR-27a and miR-24-2) at loci 19p13.13. The upregulation of these miRNAs contributed to gastric cancer progression. Additionally, the group also revealed that miR-23a downregulated MT2A and that inhibition of the microRNA halted tumor growth [6].
It was also reported that amplification of miR-191/425 promotes the growth of breast cancer [7]. The group reported that the copy number of miR-191/425 was significantly higher in primary breast tumor in comparison to normal tissue. MicroRNA-191/425 target DICER1 which leads to the global miRNA dysregulation due to its function in processing pre-miRNA into mature miRNA, thus effectively leading to the promotion of breast cancer tumorigenesis.
2. Mutation (Single Point Mutation, Deletion, Insertion, Base Substitution)
Mutation to genes such as substitution or deletion often leads to the production of nonfunctional translated proteins due to impaired codon arrangements [8][9][10]. As a consequence, critical cellular signaling pathways are frequently dysregulated due to the impairment in protein structure and function where this culminates with the development of various cancers [11][12][13][14]. A similar phenomenon can be observed in miRNA where a mutation in the miRNA locus would lead to dysfunctional miRNA expression (Figure 2).
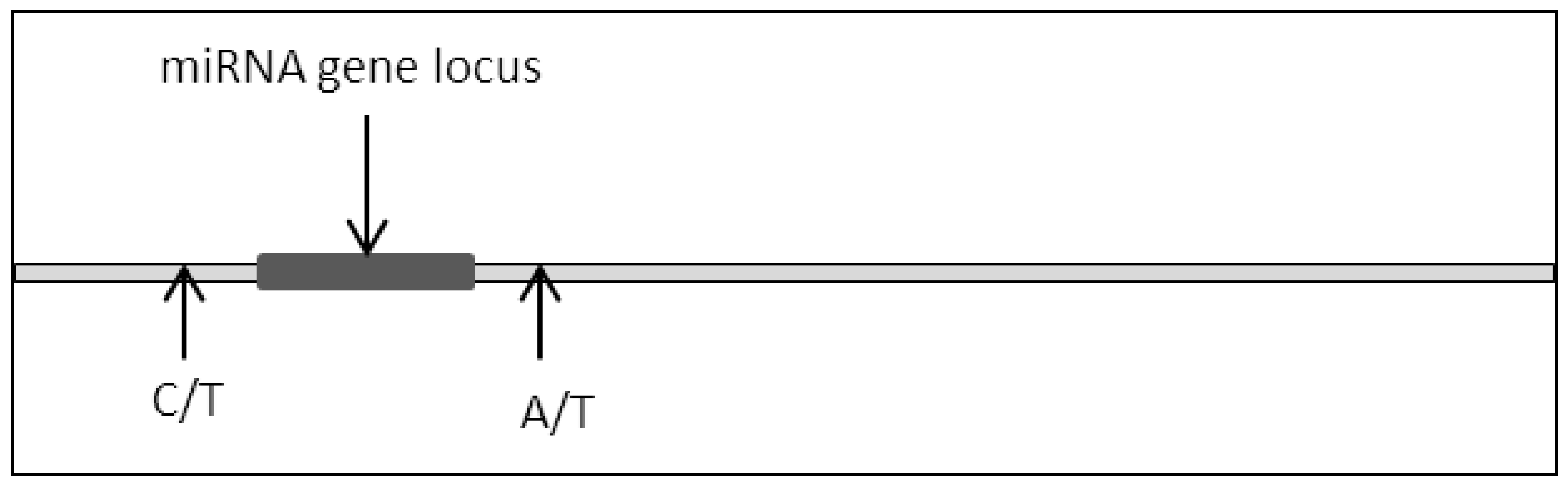
Figure 2. Single base disposition at miRNA locus.
This was proven in the case of the miR-15a and miR-16-1 locus where partial homozygous deletion of the locus led to the depletion of miR-15a expression. Furthermore, the group also reported a single base substitution of T to C 21bp upstream of the miR-15a coding sequence. It was postulated from this research that these mutations lead to the development of prostate cancer due to the prevalence of these mutations in xenografts [15].
Furthermore, point mutation of miRNA was also found to play a hand in leukemogenesis. It was found that point mutation in the miR-142 resulted in the loss of function of that miRNA. This potentiates AML towards myeloid lineages while suppressing lymphoid lineages differentiation. The loss of miR-142 leads to the accumulation of ASHL1 resulting in the maintenance of the HOXA genes. This caused the myeloid progenitors to be locked in the myeloid stage thus preventing proper cellular differentiation [16]. Additionally, it was also reported from cancer cohort (TCGA) data that the miR-142-3p seed region harbors the mutation at the 7th and 6th nucleotide where A > G and C > G/T occurs predominantly in diffuse large b-cell lymphoma [17]. Earlier results reported also that mutation in the miR-142-3p seed sequence shifted miR-142 targeting from RAC1 towards ZEB2 in diffuse large b-cell lymphoma which could attribute to disease progression [18].
3. Epigenetic Modifications on miRNA Loci
Epigenetic modulation plays a critical role in the regulation of miRNA expression [19][20]. Epigenetic markers such as DNA methylation, histone state, an abundance of CpG sites and binding of epigenetic modulators influence the accessibility of miRNA loci to various transcription machinery (Figure 3A,B). In this case, a group reported that PPARγ binds consensus sequence located on the miR-122 promoter to promote expression by inducing histone acetylation. They reported that induction of hepatocellular carcinoma cells with 5′aza-2′deoxycytidine (a compound that inhibits methyltransferase) prevented the N-cor/SMRT/HMeT complex, which is a miR-122 inhibitor from binding onto the consensus promoter sequence which in turn caused a reduction in H3K9 demethylation (heterochromatin mark on the genome, usually found to be abundant in repressed genes) and provided an accessible miR-122 locus which allowed PPARγ to bind predominantly [21].
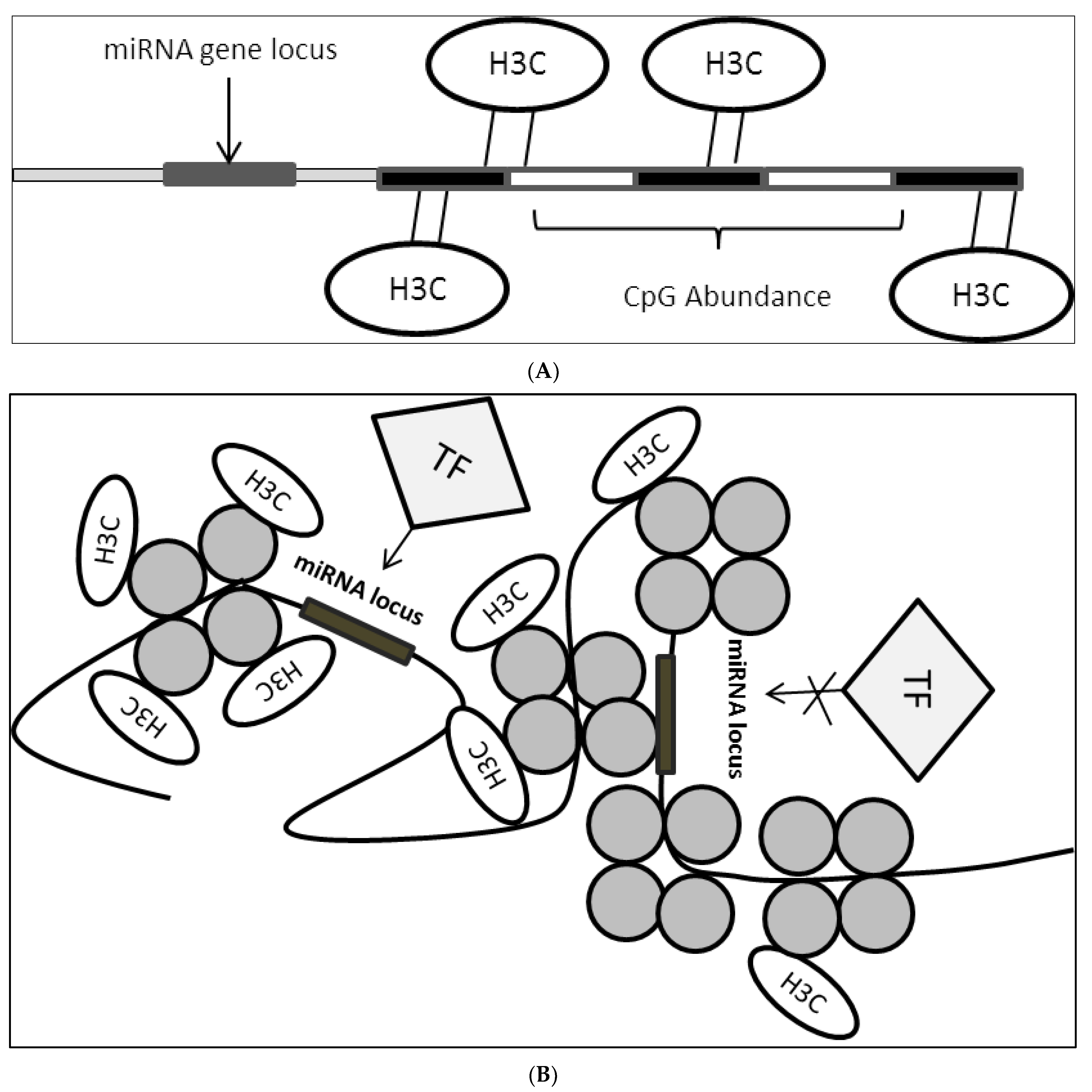
Figure 3. (A): CpG site enrichment on miRNA locus resulting in miRNA suppression. CpG methylation brought about by DNMTs methylates DNA sequence in proximity to miRNA locus or direct locus methylation leads to suppression of miRNA expression. H3C: methyl group; (B): Histone state on miRNA locus controls accessibility of TFs. Histone modification leads to the formation of open or closed chromatin structure which in turn affect TFs binding onto miRNA locus where this would result in either miRNA activation or suppression depending on the transcriptional activity of the bounded factors. H3C: methyl group.
Histone state plays a major role in the accessibility of crucial transcription machinery onto gene locus which triggers transcription or suppression. Depending on the state of chromatin brought about by the actions of histone modifiers genes can either be activated or inactivated. In miRNA regulation, it was reported that the interplay between miRNA and histone modifiers exists in modulating disease progression [22][23][24]. It was found that miR-125a-5p expression negatively regulates HDAC5 in breast cancer which contributes to disease progression via a feedback loop. The study found that miR-125a-5p binds the 3′-UTR of HDAC5 which leads to the reduction in HDAC5 dependent RUNX3 acetylation on p300/RUNX3 complex resulting in the dissociation of the complex binding on the miR-125a-5p locus. Using HDAC inhibitor, HDAC5 activity was suppressed leading to the reduction of miR-125a-5p which leads to apoptosis suggesting interplay between miRNA and HDACs in mediating apoptotic evasion [25]. In addition, it was also reported that the EZH2 histone modifier, which induces heterochromatinization, was overexpressed in pancreatic duct adenocarcinoma (PDAC) leading to the suppression of miR-218. Direct EZH2 siRNA targeting leads to restoration in miR-218 expression as this abrogates EZH2 binding onto the miRNA locus also blocking the recruitment of DNMTs via EZH2 [26].
Additionally, it was also shown that CpG sites, influence the expression of miR-335 located within the MEST gene locus downstream of MEST exon 1 which may contribute to hepatocarcinogenesis. It was found that the CpG-rich site located upstream of the miR-335 beyond exon 1 of MEST resulted in the repression of miR-335 expression due to DNA hypermethylation. The group also reveals that treatment with 5′aza-2′deoxycytidine, a known DNA demethylator, resulted in the upregulation of the suppressed miR-335 [27].
It was found that in prostate cancer overexpression of the MED1 gene would lead to the disease progression [28][29]. This effect could be mitigated via MED1 suppression by miR-205. However, it was found that the miR-205 locus was hypermethylated in prostate cancer thus leading to the epigenetic silencing of the miRNA. Additionally, the same group found that H3K9Ac, a histone mark for euchromatin (highly accessible areas within the genome usually susceptible to transcription factor binding), was significantly reduced on miR-205 loci. Furthermore, they also reported the prevalence of epigenetic in various cancer cells and found that miR-205 expression was inversely correlated with DNA methylation [30][31].
Results have shown that miR-195 was epigenetically influenced as well. It was found that the expression of miR-195, a tumor suppressor miRNA that is required for normal colon function, was downregulated in colorectal cancer cells [32][33]. It was also shown in the study that CpG islands upstream of the miR-195 locus were partially methylated in normal colorectal cells. However, in colorectal cancer, this site was fully methylated [34]. The findings from these studies suggest that DNA hypermethylation of miRNA loci in colorectal cancer plays a crucial role in oncogenesis by interfering with the tumor-suppressive activity of miR-195.
Reports have also shown that miR-340 is implicated epigenetically in neuroblastoma [35][36]. CpG-rich sites were found downstream of the predicted transcription start site (TSS) of miR-340 and these sites were extensively methylated. Interestingly, these sites were also found to overlap with RNA polymerase II (pol II) binding sites thus suggesting epigenetic modulation in transcriptional control of miRNA [37]. Treatment with 5′aza-2′deoxycytidine reduced methylation of the CpG sites which in turn led to an increase in miR-340 activity via reduction of its target gene SOX2. These findings clearly highlight the role of epigenetic modifications in miRNA dysregulation.
4. Transcription Factors (TF) Controlling miRNA Expression Dysregulation
Transcription factors (TFs) are well known to either promote or suppress gene expression [38]. Transcription factors are generally understood as proteins that bind specific consensus sequences located on genomic regions, particularly, on gene loci [39][40]. The binding of TFs can either occur proximal or distal from the TSS [41][42]. TFs that promote accessibility on the genomic region for RNA pol II binding and other TFs are known as activators whereas suppressors are TFs that promote inaccessibility [43]. The mechanism of action of TFs involves the recruitment of other TFs or epigenetic modulators on specific genomic loci. Depending on what type of TF bounded (Suppressors or Activators) they partner with different epigenome modulators (HDAC, HMET, HAT, p300). This leads to either upregulation or downregulation of genes affected [44][45].
Proper TF function is crucial for the regulation of miRNA expression. This can clearly be seen in the interaction between the transcription factor ETS5 and miR-155. It was shown that ETS5 binds to miR-155 TSS and activates the expression of the pri-miR-155 which in turn leads to lipopolysaccharide activation in the human embryonic kidney. Contrarily, the expression of miR-155 was downregulated by the addition of IL-10, an inhibitor of ETS5. This is an example of an activator mechanism in miRNA expression [46].
Additionally, it was also found that GATA3 modulates the activity of miR-29b by binding on the promoter of miR-29b [47]. Reporter assay also showed induction in expression when miR-29b loci containing the GATA3 binding sequences were cloned into luciferase plasmids [48]. However, deletion of the promoter binding sequence resulted in the reduction in luciferase activity clearly showing that GATA3 binding is crucial for miR-29b induction.
Interestingly, it was also reported that miR-1 and miR-206 were both regulated by NRF2 transcription factors in an HDAC4-dependent manner [49][50]. Downregulation of NRF2 resulted in an increase in miR-1 and miR-206 expression. This indicates that the TF acts as a suppressor. NRF induces HDAC activity resulting in a reduction in accessibility (heterochromatin) on the genomic loci, thus inactivating transcription. Furthermore, downregulation of HDAC resulted in an increase in miR-1 and miR-206 expression.
A summary of the various mechanisms involved in miRNA dysregulation is presented in Table 1.
Table 1. Table showing various mechanisms of dysregulation of miRNA in cancers.
| miRNA | Mechanism of Dysregulation | Consequence | References |
|---|---|---|---|
| miR-650 | Loci amplification | Inverse correlation was observed between miR-650 and tumour supressor genes ING4 and NDRG2 | [51] |
| miR-21 | Loci amplification | miR-21 overexpression leads to PTEN suppression | [52] |
| miR-4288 | Deletion | Loss of miRNA in prostate cancer, miRNA directly represses metastatic/invasion genes MMP16 and ROCK1 | [53] |
| miR-3613 | Deletion | miR-3613 was found to be lower in breast cancer. Gain of function reveals miR-3613 to regulate PAFAH1B2 and PDK3 blocking oncogenesis | [54] |
| miR-379 | Base substitution | Hotspot mutation commonly occurring in lung adenocarcinoma | [55] |
| miR-142-3p | Epigenetic suppression via DNMT recruitment | Hypermethylation of miR-142 leads to unfavorable prognosis in nasopharyngeal carcinoma | [56] |
| miR-338-5p/421 | EZH2 mediated suppression via DNA methylation | Presence of CpG marks on primary prostate cancer. Ectopic expression reveals suppression in prostate cancer growth | [57] |
| miR-17-92/106b-25 | CMYC driven | CMYC drive the expression of these miRNA clusters, inhibition of cMYC activators resulted in suppression of these clusters in hepatocellular carcinoma | [58] |
| miR-122 | CMYC driven | CMYC oncogene overexpression in hepatocellular carcinoma activates miR-122 via direct promoter binding driving oncogenesis | [59] |
| miR-455-3p | Reside in host gene driven by p53 | miRNA involves in cancer quiescence via p53 mediation | [60] |
References
- Lango-Chavarría, M.; Chimal-Ramírez, G.K.; Ruiz-Tachiquín, M.E.; Espinoza-Sánchez, N.A.; Suárez-Arriaga, M.C.; Fuentes-Pananá, E.M. A 22q11.2 amplification in the region encoding microRNA-650 correlates with the epithelial to mesenchymal transition in breast cancer primary cultures of Mexican patients. Int. J. Oncol. 2017, 50, 432–440.
- Hirata, Y.; Murai, N.; Yanaihara, N.; Saito, M.; Saito, M.; Urashima, M.; Murakami, Y.; Matsufuji, S.; Okamoto, A. MicroRNA-21 is a candidate driver gene for 17q23-25 amplification in ovarian clear cell carcinoma. BMC Cancer 2014, 14, 799.
- Bhagirath, D.; Yang, T.L.; Tabatabai, Z.L.; Shahryari, V.; Majid, S.; Dahiya, R.; Tanaka, Y.; Saini, S. Role of a novel race-related tumor suppressor microRNA located in frequently deleted chromosomal locus 8p21 in prostate cancer progression. Carcinogenesis 2019, 40, 633–642.
- Chen, C.; Pan, Y.; Bai, L.; Chen, H.; Duan, Z.; Si, Q.; Zhu, R.; Chuang, T.-H.; Luo, Y. MicroRNA-3613-3p functions as a tumor suppressor and represents a novel therapeutic target in breast cancer. Breast Cancer Res. 2021, 23, 12.
- Galka-Marciniak, P.; Urbanek-Trzeciak, M.O.; Nawrocka, P.M.; Dutkiewicz, A.; Giefing, M.; Lewandowska, M.A.; Kozlowski, P. Somatic Mutations in miRNA Genes in Lung Cancer—Potential Functional Consequences of Non-Coding Sequence Variants. Cancers 2019, 11, 793.
- Li, Y.; He, Q.; Wen, X.; Hong, X.; Yang, X.; Tang, X.; Zhang, P.; Lei, Y.; Sun, Y.; Zhang, J.; et al. EZH2-DNMT1-mediated epigenetic silencing of miR-142-3p promotes metastasis through targeting ZEB2 in nasopharyngeal carcinoma. Cell Death Differ. 2019, 26, 1089–1106.
- Bhatia, V.; Yadav, A.; Tiwari, R.; Nigam, S.; Goel, S.; Carskadon, S.; Gupta, N.; Goel, A.; Palanisamy, N.; Ateeq, B. Epigenetic Silencing of miRNA-338-5p and miRNA-421 Drives SPINK1-Positive Prostate Cancer. Clin. Cancer Res. 2019, 25, 2755–2768.
- Li, S.-G.; Shi, Q.-W.; Yuan, L.; Qin, L.; Wang, Y.; Miao, Y.-Q.; Chen, Z.; Ling, C.-Q.; Qin, W. C-Myc-dependent repression of two oncogenic miRNA clusters contributes to triptolide-induced cell death in hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 2018, 37, 51.
- Wang, B.; Hsu, S.; Wang, X.; Kutay, H.; Bid, H.K.; Yu, J.; Ganju, R.K.; Jacob, S.T.; Yuneva, M.; Ghoshal, K. Reciprocal regulation of microRNA-122 and c-Myc in hepatocellular cancer: Role of E2F1 and transcription factor dimerization partner 2. Hepatology 2014, 59, 555–566.
- La, T.; Liu, G.Z.; Farrelly, M.; Cole, N.; Feng, Y.C.; Zhang, Y.Y.; Sherwin, S.K.; Yari, H.; Tabatabaee, H.; Yan, X.G.; et al. A p53-Responsive miRNA Network Promotes Cancer Cell Quiescence. Cancer Res. 2018, 78, 6666–6679.
- Elli, F.M.; Ghirardello, S.; Giavoli, C.; Gangi, S.; Dioni, L.; Crippa, M.; Finelli, P.; Bergamaschi, S.; Mosca, F.; Spada, A. A new structural rearrangement associated to Wolfram syndrome in a child with a partial phenotype. Gene 2012, 509, 168–172.
- Lu, X.; Cen, Z.; Xie, F.; Ouyang, Z.; Zhang, B.; Zhao, G.; Luo, W. Genetic analysis of SPG4 and SPG3A genes in a cohort of Chinese patients with hereditary spastic paraplegia. J. Neurol. Sci. 2014, 347, 368–371.
- Ou, S.-H.I.; Azada, M.; Hsiang, D.J.; Herman, J.M.; Kain, T.S.; Siwak-Tapp, C.; Casey, C.; He, J.; Ali, S.M.; Klempner, S.J. Next-generation sequencing reveals a Novel NSCLC ALK F1174V mutation and confirms ALK G1202R mutation confers high-level resistance to alectinib (CH5424802/RO5424802) in ALK-rearranged NSCLC patients who progressed on crizotinib. J. Thorac. Oncol. 2014, 9, 549–553.
- Abe, A.; Yamamoto, Y.; Katsumi, A.; Okamoto, A.; Tokuda, M.; Inaguma, Y.; Yamamoto, K.; Yanada, M.; Kanie, T.; Tomita, A. Rearrangement of VPS13B, a causative gene of Cohen syndrome, in a case of RUNX1–RUNX1T1 leukemia with t (8; 12; 21). Int. J. Hematol. 2018, 108, 208–212.
- Porkka, K.P.; Ogg, E.-L.; Saramäki, O.R.; Vessella, R.L.; Pukkila, H.; Lähdesmäki, H.; Weerden, W.M.; van Wolf, M.; Kallioniemi, O.P.; Jenster, G.; et al. The miR-15a-miR-16-1 locus is homozygously deleted in a subset of prostate cancers. Genes Chromosomes Cancer 2011, 50, 499–509.
- Trissal, M.C.; Wong, T.N.; Yao, J.-C.; Ramaswamy, R.; Kuo, I.; Baty, J.; Sun, Y.; Jih, G.; Parikh, N.; Berrien-Elliott, M.M.; et al. MIR142 Loss-of-Function Mutations Derepress ASH1L to Increase HOXA Gene Expression and Promote Leukemogenesis. Cancer Res. 2018, 78, 3510–3521.
- Urbanek-Trzeciak, M.O.; Galka-Marciniak, P.; Nawrocka, P.M.; Kowal, E.; Szwec, S.; Giefing, M.; Kozlowski, P. Pan-cancer analysis of somatic mutations in miRNA genes. EBioMedicine 2020, 61, 103051.
- Kwanhian, W.; Lenze, D.; Alles, J.; Motsch, N.; Barth, S.; Döll, C.; Imig, J.; Hummel, M.; Tinguely, M.; Trivedi, P.; et al. MicroRNA-142 is mutated in about 20% of diffuse large B-cell lymphoma. Cancer Med. 2012, 1, 141–155.
- Meng, F.; Wehbe-Janek, H.; Henson, R.; Smith, H.; Patel, T. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene 2008, 27, 378.
- Iorio, M.V.; Piovan, C.; Croce, C.M. Interplay between microRNAs and the epigenetic machinery: An intricate network. Biochim. Biophys. Acta BBA-Gene Regul. Mech. 2010, 1799, 694–701.
- Song, K.; Han, C.; Zhang, J.; Lu, D.; Dash, S.; Feitelson, M.; Lim, K.; Wu, T. Epigenetic regulation of miR-122 by PPARγ and hepatitis B virus X protein in hepatocellular carcinoma cells. Hepatology 2013, 58, 1681–1692.
- Bourassa, M.W.; Ratan, R.R. The interplay between microRNAs and histone deacetylases in neurological diseases. Neurochem. Int. 2014, 77, 33–39.
- Tomasetti, M.; Gaetani, S.; Monaco, F.; Neuzil, J.; Santarelli, L. Epigenetic Regulation of miRNA Expression in Malignant Mesothelioma: miRNAs as Biomarkers of Early Diagnosis and Therapy. Front. Oncol. 2019, 9, 1293.
- Li, G.; Tian, Y.; Zhu, W.-G. The Roles of Histone Deacetylases and Their Inhibitors in Cancer Therapy. Front. Cell Dev. Biol. 2020, 8, 1004.
- Hsieh, T.-H.; Hsu, C.-Y.; Tsai, C.-F.; Long, C.-Y.; Wu, C.-H.; Wu, D.-C.; Lee, J.-N.; Chang, W.-C.; Tsai, E.-M. HDAC Inhibitors Target HDAC5, Upregulate MicroRNA-125a-5p, and Induce Apoptosis in Breast Cancer Cells. Mol. Ther. 2015, 23, 656–666.
- Li, C.H.; To, K.; Tong, J.H.; Xiao, Z.; Xia, T.; Lai, P.B.S.; Chow, S.C.; Zhu, Y.; Chan, S.L.; Marquez, V.E.; et al. Enhancer of Zeste Homolog 2 Silences MicroRNA-218 in Human Pancreatic Ductal Adenocarcinoma Cells by Inducing Formation of Heterochromatin. Gastroenterology 2013, 144, 1086–1097.
- Dohi, O.; Yasui, K.; Gen, Y.; Takada, H.; Endo, M.; Tsuji, K.; Konishi, C.; Yamada, N.; Mitsuyoshi, H.; Yagi, N. Epigenetic silencing of miR-335 and its host gene MEST in hepatocellular carcinoma. Int. J. Oncol. 2013, 42, 411–418.
- Srivastava, A.; Goldberger, H.; Dimtchev, A.; Ramalinga, M.; Chijioke, J.; Marian, C.; Oermann, E.K.; Uhm, S.; Kim, J.S.; Chen, L.N. MicroRNA profiling in prostate cancer-the diagnostic potential of urinary miR-205 and miR-214. PLoS ONE 2013, 8, e76994.
- Gandellini, P.; Giannoni, E.; Casamichele, A.; Taddei, M.L.; Callari, M.; Piovan, C.; Valdagni, R.; Pierotti, M.A.; Zaffaroni, N.; Chiarugi, P. miR-205 hinders the malignant interplay between prostate cancer cells and associated fibroblasts. Antioxid. Redox Signal 2014, 20, 1045–1059.
- Hulf, T.; Sibbritt, T.; Wiklund, E.D.; Patterson, K.; Song, J.Z.; Stirzaker, C.; Qu, W.; Nair, S.; Horvath, L.G.; Armstrong, N.J.; et al. Epigenetic-induced repression of microRNA-205 is associated with MED1 activation and a poorer prognosis in localized prostate cancer. Oncogene 2013, 32, 2891–2899.
- Babu, A.; Verma, R.S. Chromosome structure: Euchromatin and heterochromatin. In International Review of Cytology; Elsevier: Amsterdam, The Netherlands, 1987; Volume 108, pp. 1–60.
- Wang, X.; Wang, J.; Ma, H.; Zhang, J.; Zhou, X. Downregulation of miR-195 correlates with lymph node metastasis and poor prognosis in colorectal cancer. Med. Oncol. 2012, 29, 919–927.
- Liu, L.; Chen, L.; Xu, Y.; Li, R.; Du, X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem. Biophys. Res. Commun. 2010, 400, 236–240.
- Menigatti, M.; Staiano, T.; Manser, C.N.; Bauerfeind, P.; Komljenovic, A.; Robinson, M.; Jiricny, J.; Buffoli, F.; Marra, G. Epigenetic silencing of monoallelically methylated miRNA loci in precancerous colorectal lesions. Oncogenesis 2013, 2, e56.
- Domingo-Fernandez, R.; Watters, K.; Piskareva, O.; Stallings, R.L.; Bray, I. The role of genetic and epigenetic alterations in neuroblastoma disease pathogenesis. Pediatr. Surg. Int. 2013, 29, 101–119.
- Ram Kumar, R.M.; Schor, N.F. Methylation of DNA and chromatin as a mechanism of oncogenesis and therapeutic target in neuroblastoma. Oncotarget 2018, 9, 22184–22193.
- Das, S.; Bryan, K.; Buckley, P.G.; Piskareva, O.; Bray, I.M.; Foley, N.; Ryan, J.; Lynch, J.; Creevey, L.; Fay, J.; et al. Modulation of neuroblastoma disease pathogenesis by an extensive network of epigenetically regulated microRNAs. Oncogene 2013, 32, 2927–2936.
- Harbison, C.T.; Gordon, D.B.; Lee, T.I.; Rinaldi, N.J.; Macisaac, K.D.; Danford, T.W.; Hannett, N.M.; Tagne, J.-B.; Reynolds, D.B.; Yoo, J. Transcriptional regulatory code of a eukaryotic genome. Nature 2004, 431, 99.
- Wingender, E.; Dietze, P.; Karas, H.; Knüppel, R. TRANSFAC: A database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996, 24, 238–241.
- Cawley, S.; Bekiranov, S.; Ng, H.H.; Kapranov, P.; Sekinger, E.A.; Kampa, D.; Piccolboni, A.; Sementchenko, V.; Cheng, J.; Williams, A.J. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 2004, 116, 499–509.
- Stadler, M.B.; Murr, R.; Burger, L.; Ivanek, R.; Lienert, F.; Schöler, A.; van Nimwegen, E.; Wirbelauer, C.; Oakeley, E.J.; Gaidatzis, D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011, 480, 490–495.
- Spitz, F.; Furlong, E.E. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613.
- Pei, L.; He, X.; Li, S.; Sun, R.; Xiang, Q.; Ren, G.; Xiang, T. KRAB zinc-finger protein 382 regulates epithelial-mesenchymal transition and functions as a tumor suppressor, but is silenced by CpG methylation in gastric cancer. Int. J. Oncol. 2018, 53, 961–972.
- Puisieux, A.; Brabletz, T.; Caramel, J. Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 2014, 16, 488.
- Shu, X.-Z.; Zhang, L.-N.; Zhang, R.; Zhang, C.-J.; He, H.-P.; Zhou, H.; Wang, N.; Zhang, T.-C. Histone acetyltransferase p300 promotes MRTF-A-mediates transactivation of VE-cadherin gene in human umbilical vein endothelial cells. Gene 2015, 563, 17–23.
- Quinn, S.R.; Mangan, N.E.; Caffrey, B.E.; Gantier, M.P.; Williams, B.R.; Hertzog, P.J.; McCoy, C.E.; O’Neill, L.A. The role of Ets2 transcription factor in the induction of microRNA-155 (miR-155) by lipopolysaccharide and its targeting by interleukin-10. J. Biol. Chem. 2014, 289, 4316–4325.
- Liu, H.; Wang, B.; Lin, J.; Zhao, L. microRNA-29b: An emerging player in human cancer. Asian Pac. J. Cancer Prev. 2014, 15, 9059–9064.
- Zhang, K.; Cai, H.-X.; Gao, S.; Yang, G.-L.; Deng, H.-T.; Xu, G.-C.; Han, J.; Zhang, Q.-Z.; Li, L.-Y. TNFSF15 suppresses VEGF production in endothelial cells by stimulating miR-29b expression via activation of JNK-GATA3 signals. Oncotarget 2016, 7, 69436–69449.
- Wang, S.-H.; Zhang, W.-J.; Wu, X.-C.; Zhang, M.-D.; Weng, M.-Z.; Zhou, D.; Wang, J.-D.; Quan, Z.-W. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget 2016, 7, 37857.
- Han, C.; Shen, J.K.; Hornicek, F.J.; Kan, Q.; Duan, Z. Regulation of microRNA-1 (miR-1) expression in human cancer. Biochim. Biophys. Acta BBA-Gene Regul. Mech. 2017, 1860, 227–232.
More
Information
Subjects:
Oncology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
698
Revisions:
4 times
(View History)
Update Date:
05 May 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




