Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | ANDRÁS Habil FODOR | -- | 2890 | 2022-04-15 02:07:46 | | | |
| 2 | Conner Chen | -28 word(s) | 2862 | 2022-04-15 02:50:55 | | | | |
| 3 | Conner Chen | -8 word(s) | 2854 | 2022-04-20 05:59:38 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Fodor, A. The Prokaryotic Partner of Entomopathogenic-Nematode/Bacterium Symbiotic Associations. Encyclopedia. Available online: https://encyclopedia.pub/entry/21803 (accessed on 07 February 2026).
Fodor A. The Prokaryotic Partner of Entomopathogenic-Nematode/Bacterium Symbiotic Associations. Encyclopedia. Available at: https://encyclopedia.pub/entry/21803. Accessed February 07, 2026.
Fodor, András. "The Prokaryotic Partner of Entomopathogenic-Nematode/Bacterium Symbiotic Associations" Encyclopedia, https://encyclopedia.pub/entry/21803 (accessed February 07, 2026).
Fodor, A. (2022, April 15). The Prokaryotic Partner of Entomopathogenic-Nematode/Bacterium Symbiotic Associations. In Encyclopedia. https://encyclopedia.pub/entry/21803
Fodor, András. "The Prokaryotic Partner of Entomopathogenic-Nematode/Bacterium Symbiotic Associations." Encyclopedia. Web. 15 April, 2022.
Copy Citation
The natural role of peptide-antimicrobials, produced by the prokaryotic partner of entomopathogenic-nematode/bacterium (EPN/EPB) symbiotic associations, is to sustain monoxenic conditions for the EPB in the gut of the semi-anabiotic infective dauer juvenile (IJ) EPN. They keep pathobiome conditions balanced for the EPN/EPB complex in polyxenic (soil, vanquished insect cadaver) niches. Xenorhabdus szentirmaii DSM16338(T) (EMC), and X. budapestensis DSM16342(T) (EMA), are the respective natural symbionts of EPN species Steinernema rarum and S. bicornutum.
Xenorhabdus 1
EPN/EPB cospeciation 10
1. Coevolution and Co-Speciation of EPN/EPB Symbiotic Associations
Except for the human pathogenic Photorhabdus asymbiotica [1][2][3], no EPB bacteria can be found in the soil as a free-living organism, but only in the colonized insect cadavers, and the monoxenically colonized gut of the infective dauer juvenile (IJ) developmental variant EPB [4], as symbiotic partners of the respective EPN. Many EPN/EPB associations have been discovered so far. Two EPN genera (Steinernema, Heterorhabditis), and 2 EPB genera (Xenorhabdus, Photorhabdus) are involved. Each EPN and EPB genus includes several species, subspecies, and strains.
Each Steinernema EPN strain is capable of establishing symbiosis with one or more, but a very limited number of, Xenorhabdus strains, and exclusively with Xenorhabdus, with no exception [5], which usually, but not exclusively, belong to the same species or subspecies.
Each Heterorhabditis EPN strain is capable of establishing symbiosis with one or more, but a very limited, number of Photorhabdus strains, but exclusively with Photorhabdus, with no exception [4]), usually, but not exclusively, belonging to the same species or subspecies. In the case of EPB species, the rDNA sequence-based subclusters [6][7], more-or-less correspond to subspecies rank [8][9]), as demonstrated in (Figure 1) [8][9][10][11][12][13][14][15][16].
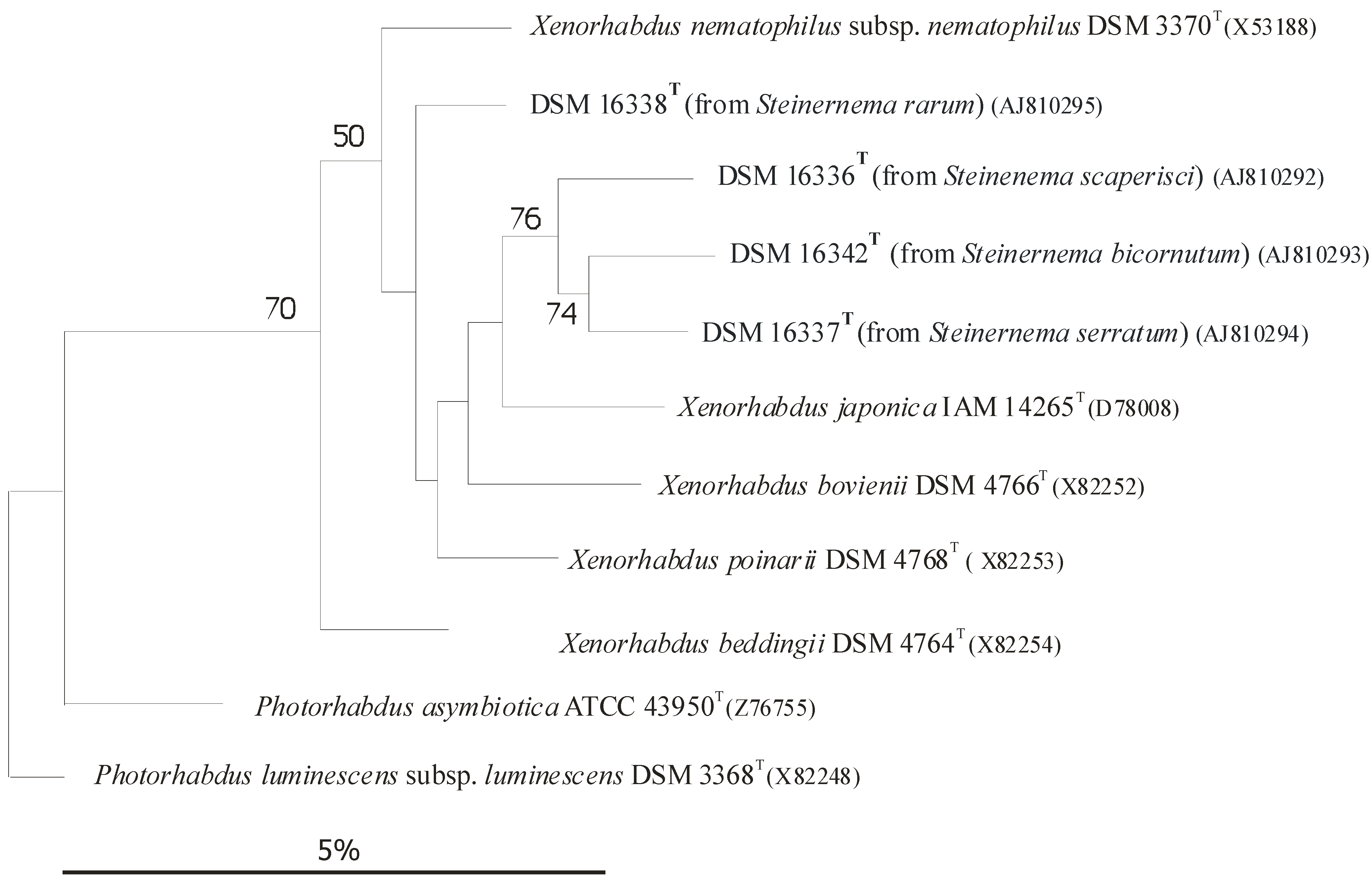
Figure 1. Dendrogram of 16S rRNA gene sequence similarities of Xenorhabdus species generated by distance matrix analysis. Dendrogram of 16S rRNA gene sequence similarities generated by distance matrix analysis. Figure 1 demonstrates the close taxonomic relation between Xenorhabdus and Photorhabdus genera.
2. Coevolution via Co-Speciation: Antimicrobial Active Peptides as Strategic Weapons Used in the Struggle to Conquest a Given Niche
In a given niche there are usually more than one EPN/EPB symbiotic complexes present and competing with each other if their insect targets are the same. The coevolution of interacting species can lead to codependent mutualists [17]. The precondition for evolutionary fixation of an EPN/EPB symbiotic complex in a given niche of a respective EPN/EPB complex is to win the struggle of insect prey against natural enemies, as well as competitors. Meanwhile, the mutualism should be kept [17]. Each symbiotic EPB (Xenorhabdus, Photorhabdus) partner owns an individual set of chemical arsenals for these unavoidable battles.
2.1. Battle with the Insect Prey Using Toxins
2.2. Battle with EPN Competitors Using Rhabdopeptides
Seven linear peptides named rhabdopeptides I-O, 1–7, were recently isolated from the cell-free culture media (CFCM) of X. budapestensis SN84 [23]. The structures of the peptides were elucidated based on extensive mass spectrometry (MS), and nuclear magnetic resonance (NMR), analyses. Rhabdopeptides I-3, rhabdopeptides I-4, and rhabdo-peptides I-7 were novel compounds. All seven compounds were tested for their nematicidal activities against the second-stage juveniles (J2) of Meloidogyne incognita. Rhabdopeptide I-2 demonstrated strong inhibitory activity [23].
2.3. Battle between Competitor EPBs Using Xenorhabdicins
Different Steinernema EPN species coexist with different Xenorhabdus symbionts when invading the same insect, setting up a competition for nutrients within the insect cadaver. The different Xenorhabdus species produce both diverse antibiotic compounds and prophage-derived R-type bacteriocins, xenorhabdicins [24]. The functions of these molecules during competition also seems extremely important from the aspect of coevolution.
Anti-Xenorhabdus activities of strains representing the 7 Xenorhabdus species against each other, and non-related Gram-negative bacteria, were compared in LB media [25]. The strongest anti-Xenorhabdus activity was shown by the CFCM of. X. bovienii NYH, (a symbiont of S. feltiae, isolated by AF in Nyíregyháza, Hungary) [26]. This showed a moderate antibacterial activity against Gram-negative bacteria Escherichia coli and Klebsiella pneumoniae (see Figure 2D), compared to other Xenorhabdus species.
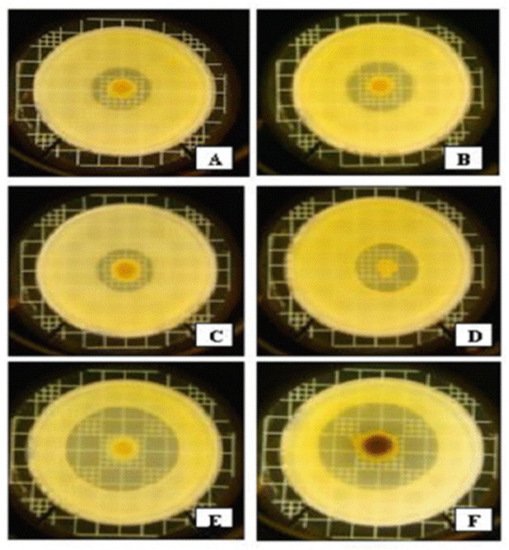
Figure 2. Interspecific differences in anti-Gram-negative activities within the genus Xenorhabdus based on overlay bioassays in LBA media on Klebsiella pneumoniae (mastitis isolates from cows). Each bacterium colony was grown from a 5 μL dropping of an overnight liquid (LB) culture on the surface of LBA medium for 5 days at 25 °C, and overlaid with 3 mL of soft (0.05 w/v) agar containing 0.3 mL log-phase (OD = 0.25) liquid (LB) culture of mastitis isolate Kl. pneumoniae in the Hogan laboratory at the Ohio State University, Wooster, OH, USA [27]. A = X. nematophila DSM3370; B = X. cabanillasii BP; C = X. nematophila ATCC 196061(T); D = X. bovienii NYH; E = X. budapestensis DSM16342(T); F = X. szentirmaii DSM16338(T), and cultured at 37 °C overnight. Note that by far the largest inactivation zones can be seen around the EMA colony (5E) and EMC (5F). Note the color of the EMC colony caused by iodinin crystals on the surface; see later).
The CFCM of X. ehlersii was also toxic to many other Xenorhabdus, but completely ineffective against E. coli OP50, or Kl. pneumoniae. On the other hand, the strongest antibiotic producers, X. budapestensis and X. szentirmaii (Figure 2E,F, respectively), were rather vulnerable to the anti-Xenorhabdus compounds produced by the others. Meanwhile, their compounds were barely effective against other Xenorhabdus species, at least on complete (LBA) media. Xenorhabdus innexi, a moderate anti-Gram-negative antibiotic producer, proved highly tolerant to the anti-Xenorhabdus compounds of others, with the exception of X. bovienii NYH [26].
The conclusion is that there was no correlation between the general anti-Gram negative and the anti-Xenorhabdus activities, but there was a positive correlation demonstrated between the anti-Xenorhabdus activities and sensitivity to anti-Xenorhabdus compounds in the CFCM [25].
10 years later in another experiment [28], using another X. bovienii strain, the natural symbiont of S. jollieti, (called Xb-Sj) was a very weak antibiotic producer. It possesses a P2-like phage tail gene cluster (xbp1), that encodes genes for xenorhabdicin production (Steven A. Forst, personal communication). Purified xenorhabdicins from the CFCM of X. bovienii Xb-Sj strain exerted a sharp, but narrow, spectrum of activity only towards Xenorhabdus and Photorhabdus species [28] (Thappeta et al., 2020).
In that experiment, X. szentirmaii was extremely sensitive towards the purified X. bovienii xenorhabdicin, and it did not produce effective xenorhabdicin against the X. bovienii Xb-Sj strain, at least not in poor Grace’s medium [28]. However, it was demonstrated that X. szentirmaii produced high-level antibiotic activity, which killed X. bovienii in a complete rich medium [28]. When the two species were co-cultured in either of the two media, X. szentirmaii was the winner. One can conclude that in nature the production of antibiotics is probably predominant in interspecies competition [29].
In the battle to win over food competitors by using AMPs and other secondary metabolites, the most successful symbiotic EPN/EPB complexes should be able to produce the best antimicrobial peptides to win against food-competitor microorganisms.
3. Antimicrobial Peptides from EMC and EMA, Fabclavines from both, and Phenazines from EMC
There have been a few biosynthetic AMP families discovered in the Xenorhabdus species over the last decade, and providing the complete inventory of them is out of the scope. Enzymes called ’non-ribosomal templalted peptide synthetizers’ (NRPSs) produce a wide variety of different natural peptid products from amino acid precursors [29]. These non-ribosomal encoded peptides (NRPs) are of short chain lengths. The common features of these molecular families are as follows. Each of them is a hybrid molecule, enzymatically synthesized by enzymes encoded by the members of a respective biosynthetic gene cluster (BGC) consisting of cooperating genes. The corresponding biosynthetic gene clusters (BGCs) could easily be identified by gene-sequence-similarity-based bioinformatics strategies [30]. Until recently, the actual access to these biosynthetic natural products for structure elucidation and bioactivity testing had been extremely difficult. The Bode laboratory recently discovered that the global post-transcriptional regulator, Hfq, which is widespread in bacteria and performs many functions, one of which is the facilitation of sRNA binding to target mRNAs, exerts several other pleiotropic effects [31]. A complete hfq deletion mutant EPB is no longer capable of sustaining a healthy symbiosis with its EPN partner due to the abolition of the production of all known secondary metabolites [31], i.e., the deletion of the gene encoding the RNA chaperone, Hfq, results in strains losing the production of most synthetic natural products, including NRPs [32]. Each contained a non-ribosomal-templated poly-amine (NRP) moiety. Each BGC encodes for one branch of nonribosomal peptide synthetases (NRPSs) [29][33][34]. In general, the NRPS consist of polypeptides, with a unidirectional interaction order, from N-terminal to C-terminal. There are usually adenylation domains, thiolation domains, condensation domains, dual condensation/epimerization domains, and thioesterase domains, involved (see [29], Supplementary Figure).
3.1. The Most Potent NRP-AMP Families of Xenorhabdus Origin
The Lysine-Rich, Cyclo-Lipopeptide, Molecular family
This family was discovered in X. nematophila by M. Gualtieri, and his associates [30]. It is also called Peptide Antimicrobial and is of the Xenorhabdus species (PAX peptides is the name introduced by Thaler and the other members of that research team). The biosynthesis pathway of lysine-rich cyclic peptides in X. nematophila was made by the Bode team in Frankfurt, Germany [35].
The fabclavine molecular family
This extremely important molecular family was discovered in EMA (the type-strain of X. budapestensis), and iyd its biosynthesis pathway was discovered in EMC (the type-strain of X. szentirmaii). Fabclavine [36] was identified as a bioactive, non-ribosomal encoded (NRP) peptide-polyketide-polyamine hybrid [37]. As revealed by detailed NMR and MS methods, the fabclavine analogs are hybrid secondary metabolites derived from nonribosomal peptide synthetases (NRPS) and polyunsaturated fatty acids (PUFA) [38], [37]. As mentioned earlier, a structural analog, nemaucin [39], of the peptidic part of fabclavine was discovered by the Gualtieri team earlier from X. cabanillasii (Patent. WO2012085177A1, Nosopharm, Nîmes, France, 2012). It was published as an antibiotic compound purified from X. cabanillasii strain CNCM I-4418 [39].
Fabclavine derivatives could also be found in almost all known Xenorhabdus species, but the details of the enzymatic biosynthesis of fabclavine were revealed in X. szentirmaii by [40]. They used deletion mutants of the gene encoding the RNA chaperone, Hfq, and then by exchanging the native promoter of the fabclavine (fcl) BGC against an inducible promoter in Δhfq mutants, (easy PACId approach, easy Promoter Activated Compound Identification technique) [32], resulting in the exclusive production of the corresponding fabclavine from the targeted BGC in X. szentirmaii [40], and later in other Xenorhabdus species [41]. Altogether, 32 members of the fabclavine family are now known [41].
The fabclavine biosynthesis in different Xenorhabdus species is catalyzed by a very similar biosynthetic enzyme complex (Peptide-Antimicrobial Xenorhabdus Protein Synthetase) coded by biosynthesis gene clusters (BGC), including enzymes needed for polyamine synthesis [41]. Most Xenorhabdus species are capable of synthesizing fabclavine analogs in a rather conservative manner, and the genetic differences in amino acid sequences of the NRPS-PKS genes cannot explain the species-differences in antimicrobial activities.
It was suggested that differential virulence of Xenorhabdus strains (demonstrated in Figure 2) must be caused by the difference in the global leucine-responsive regulatory protein expression level metabolites [42][43][44][45][46], leading to a difference in the production of indole compounds, and other NRPS-PKS-associated secondary metabolites [42].
The antimicrobial peptides which are effective against intruder competitors (belonging to different prokaryotic and eukaryotic taxa) competing for the same environmental niche, serve as a powerful toolkit for promoting local co-evolutionarily fixation [47] of the respective EPN/EPB symbiotic complex.
3.2. Iodinin and Phenazines
Xenorhabdus szentirmaii has extremely unusual phenotypes. One of them is their swarming behavior, and the other is exocrystal production [48]. Their motilities, both swimming and swarming, are much stronger than in any study published for a Photorhabdus or Xenorhabdus species [49][50][51][52][53][54].
The Exo-Crystal of EMC, and the Iodinin Biosynthesis as a Part of the Phenazine Pathway.
Basic Observation
Antibiotic pigment crystals were discovered and isolated by Máthé-Fodor in 2003, unpublished, but presented by Fodor et al. (BABE-2015 6th World Congress on Bioavailability & Bioequivalence: BA/BE Studies Summit 17–19 August 2015). An interesting phenomenon was discovered in the lab. After a few days of culturing X. szentirmaii on either NA, LBA, NBTA, or LBTA agar plates, the surfaces of the colonies became brilliant metallic red. At the same time, small crystals, as well as red colored oily drops, could be seen, first with a transmission light microscope, and later with the naked eye, both in the agar and liquid media (Figure 3).

Figure 3. Formation of iodinin exocrystals on and under colonies of antibiotic producing Xenorhabdus szentirmaii DSM16338(T) (EMC). Crystals on agar plates (left) and in liquid cultures in (API) test tubes (right). (Center), 40× magnification (Jenaval Light Microscope).
The number and size of the crystals increases day by day. Crystals closer to the colonies were larger and continuous, whereas those located farther away were smaller and dendritic in nature (Figure 3, Center). On other media, large red-colored oily drops formed at the edges of the colonies. In solid media, the number of oily drops was higher closer to the center of the colonies, and lower farther out [48][55]. It appears that cells of X. szentirmaii release a precursor material that is water-soluble and colorless. When OUTSIDE of the cells, this material changes color and becomes water-insoluble, and separates, either dissolved in oil droplets, or crystallizing on the surface and inside the agar media. This red colored material was later found to be iodinin (5,10-dioxidophenazine-5,10-diium-1,6-diol) [56]. Iodinin is a well-known, natural, phenazine dioxide, compound that was recently “rediscovered” as, among others, possessing potent and selective cytotoxic properties towards myeloid leukemia cell lines [57][58][59][60], but the water-insolubility complicates clinical application [61].
The colored oil droplets or pigment crystals form inside the agar medium, even if sterile cellophane separates the surface of the bacteria colonies from the agar. The cellophane Millipore 0.22 µm filter prevents the bacteria from passing into the agar, but iodinin still separates in the agar under those conditions. Two possible interpretations of this observation can be imagined. L. Haynes proposes that a water-soluble form of iodinin, rather than a chemically distinct precursor, could have been released by the cells. He proposes that the iodinin might be complexed by a water-soluble carbohydrate, which makes the complex water-soluble and gives it the ability to pass through the cellophane Millipore filter. Once in the agar, the non-covalently bound partner molecules separate, and the much less water soluble iodinin takes the form of either oil droplets or crystals. An alternative idea is that there is a water-soluble precursor, chemically distinct from iodinin, which is released by the cells and is able to pass through the cellophane and accumulate in the agar medium. In the medium it undergoes a condensation reaction to form iodinin, either spontaneously and not enzymatically, or by the catalytic action of an exo-enzyme released by the bacterium. The much less water soluble iodinin then separates from the aqueous medium as either an oil or as crystals.
Identification of the Material as Iodinin
Crystals were finally isolated using a double layer of sterile cellophane covering an LB plate, and over-layered with a bacterium suspension. Using single crystal X-ray diffraction, the pigment crystal was identified by Haynes and Zeller as iodinin [48], (see also in Figure 4). Details of the structure determination are given in Appendix C and Figure 4. For references see [62][63][64][65][66][67][68][69].
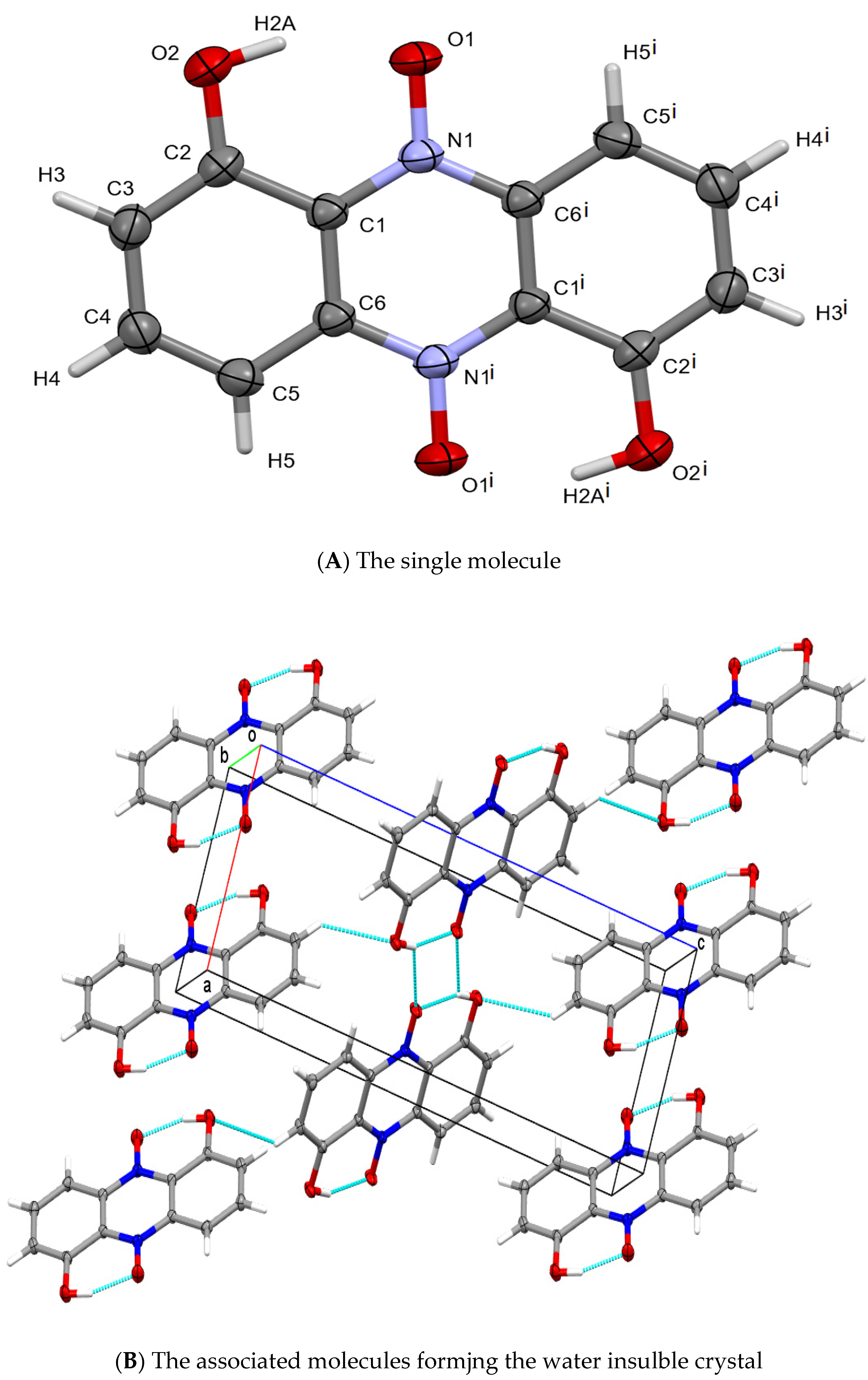
Figure 4. Representations of the chemical and crystallographic structure of iodinin. Legend: The chemical structure of the colored component (iodinin) of the exocrystal produced by X. szentirmaii. (A) the single crystal structure of iodinin. Crystallographic parameters: Monoclinic, P21/c: a = 6.0298(5), b = 5.0752(4), c = 15.854(1) Å, b = 90.421(2)°. Crystal size: 0.48 × 0.15 × 0.02 mm. θ range: 2.57 to 28.28°. Data/restraints/parameters: 1206/0/83. GooF: 1.178. R values [I > 2σ(I)]: R1 = 0.0699, wR2 = 0.1659. (B) packing of iodinin in the solid-state is dominated by π-stacked layers connected by C-H⋅⋅⋅O intercations making it largely insolubile in water. The structure of iodinin (from another organism) was previously reported [56].
Crystal Mutants in X. szentirmaii DSM16338T (EMC)
Fodor carried out Tn-mutagenesis experiments, screening for exocrystal-minus mutants. A total of 22 anti-microbial crystal mutants from X. szentirmaii were isolated. Some of these can be seen in Figure 20 in the article [55]. One mutant produced colorless oily drops (on the left side), others produced dark oils (in the right of the picture) while the wild type produced purple colored (Medium) oily drops on ENGM plates. The mutants were deposited in the stock collection of Professor Heidi Goodrich-Blair (University of Wisconsin, Madison, WI, USA). The Bode Laboratory recently discovered diversity-oriented modifications of the phenazine core through two distinct BGCs in X. szentirmaii. A previously unidentified aldehyde intermediate, which can be modified by multiple enzymatic and non-enzymatic reactions, is a common intermediate bridging the pathways encoded by the respective biosynthetic gene clusters BGCs [70].
The Discovery of a Unique Phenazine Biosynthesis Pathway in X. szentirmaii by the Bode Laboratory.
From an antiSMASH22 analysis of 28 Xenorhabdus and Photorhabdus genome sequences in the Bode Laboratory, four strains encoding phenazine BGC(s) were identified, but only X. szentirmaii from laboratory encoded two phenazine BGCs [71]. The second BGC was silent under laboratory conditions. The first includes 7 genes (A, B, C, D, E, T, F) with the same transcription orientation (5′–3′) as for the phenazine core biosynthesis. This is followed by gene U, of unknown function, and opposite transcription, followed by gene V, of unknown function, but similar transcription (orientated as A–F), finally followed by genes G and H, encoding for iodinin biosynthesis, [71]. and maintaining the same transcription (5′–3′) orientation as A–F [70]. Although the authors specifically pointed this out, please note that in this pathway, unlike the second, no NRPS-like enzyme-coding gene is represented.
References
- Gerrard, J.; Waterfield, N.; Vohra, R.; ffrench-Constant, R. Human infection with Photorhabdus asymbiotica: An emerging bacterial pathogen. Microb. Infect. 2004, 6, 229–237.
- Hapeshi, A.; Waterfield, N.R. Photorhabdus asymbiotica as an insect and human pathogen. In Current Topics in Microbiology and Immunology; Springer: Cham, Switzerlan, 2017; Volume 402, pp. 159–177.
- Vlisidou, I.; Hapeshi, A.; Healey, J.R.; Smart, K.; Yang, G.; Waterfield, N.R. The Photorhabdus asymbiotica virulence cassettes deliver protein effectors directly into target eukaryotic cells. Elife 2019, 8, e46259.
- Forst, S.; Nealson, K. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Rev. 1996, 60, 21–43.
- Forst, S.; Dowds, B.; Boemare, N.; Stackebrandt, E. Xenorhabdus and Photorhabdus spp.: Bugs that kill bugs. Annu. Rev. Microbiol. 1997, 51, 47–72.
- Szállás, E.; Koch, C.; Fodor, A.; Burghart, J.; Buss, O.; Szentirmai, A.; Nealson, K.H.; Stackebrandt, E. Phylogenetic evidence for the taxonomic heterogeneity of Photorhabdus luminescens. Int. J. Syst. Bacteriol. 1997, 47, 402–407.
- Szállás, E.; Pukall, R.; Pamjav, H.; Kovács, G.; Buzás, Z.; Fodor, A.; Stackebrandt, E. Passengers who missed the train: Comparative sequence analysis, PhastSystem PAGE-PCR-RFLP and automated RiboPrint Phenotypes of Photorhabdus strains In Development in Entomopathogenic Nematode/Bacterial Research; Griffin, C.T., Burnell, A.M., Downes, M.J., Mulder, R., Eds.; European Commission Publications: Luxemburg, 2001; pp. 36–53.
- Tailliez, P.; Pagès, S.; Ginibre, N.; Boemare, N. New insight into diversity in the genus Xenorhabdus, including the description of ten novel species. Int. J. Syst. Evol. Microbiol. 2006, 56, 2805–2818.
- Tailliez, P.; Laroui, C.; Ginibre, N.; Paule, A.; Pagès, S.; Boemare, N. Phylogeny of Photorhabdus and Xenorhabdus based on universally conserved protein-coding sequences and implications for the taxonomy of these two genera. Proposal of new taxa: X. vietnamensis sp. nov., P. luminescens subsp. caribbeanensis subsp. nov., P. luminescens subsp. hainanensis subsp. nov., P. temperata subsp. khanii subsp. nov., P. temperata subsp. tasmaniensis subsp. nov., and the reclassification of P. luminescens subsp. thracensis as P. temperata subsp. thracensis comb. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 1921–1937.
- de Soete, G. A least square algorithm for fitting additive trees to proximity data. Psychometrika 1983, 48, 621–626.
- Munro, H.N. (Ed.) Jukes and Cantor, Evolution of protein molecules. In Mammalian Protein Metabolism; Academic Press: New York, NY, USA, 1969; pp. 21–132.
- Felsenstein, J. PHYLIP Phylogeny Inference Package, Version 3.5.1; Department of Genetics, University of Washington: Seattle, WA, USA, 1993.
- Triga, D.; Pamjav, H.; Vellai, T.; Fodor, A.; Buzás, Z. Gel electrophoretic restriction fragment length polymorphism analysis of DNA derived from individual nematodes, using the PhastSystem. Electrophoresis 1999, 20, 1274–1279.
- Pamjav, H.; Triga, D.; Buzás, Z.; Vellai, T.; Lucskai, A.; Adams, B.; Reid, A.P.; Burnell, A.; Griffin, C.; Glazer, I.; et al. Novel application of PhastSystem polyacrylamide gel electrophoresis using restriction fragment length polymorphism--internal transcribed spacer patterns of individuals for molecular identification of entomopathogenic nematodes. Electrophoresis 1999, 20, 1266–1273.
- Böszörményi, E. Entomopathogen Bacterium Antibiotic Activity and Symbiotic Capacity of Gnotobiological Analyses. Ph.D Thesis, Eötvös University, Budapest, Hungary, 2010. Available online: http://teo.elte.hu/minosites/tezis2010/burgettine_boszormenyi_e.pdf (accessed on 1 August 2019).
- Machado, R.A.R.; Wüthrich, D.; Kuhnert, P.; Arce, C.C.M.; Thönen, L.; Ruiz, C.; Zhang, X.; Robert, C.A.M.; Karimi, J.; Kamali, S.; et al. Whole-genome-based revisit of Photorhabdus phylogeny: Proposal for the elevation of most Photorhabdus subspecies to the species level and description of one novel species Photorhabdus bodei sp. nov., and one novel subspecies Photorhabdus laumondii subsp. clarkei subsp. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 2664–2681.
- Morran, L.T.; Penley, M.J.; Byrd, V.S.; Meyer, A.J.; O’Sullivan, T.S.; Bashey, F.; Goodrich-Blair, H.; Lively, C.M. Nematode-bacteria mutualism: Selection within the mutualism supersedes selection outside of the mutualism. Evolution 2016, 70, 687–695.
- Yang, J.; Zeng, H.M.; Lin, H.F.; Yang, X.F.; Liu, Z.; Guo, L.H.; Yuan, J.J.; Qiu, D.W. An insecticidal protein from Xenorhabdus budapestensis that results in prophenoloxidase activation in the wax moth, Galleria mellonella. J. Invertebr. Pathol. 2012, 110, 60–67.
- Hemalatha, D.; Prabhu, S.; Rani, W.B.; Anandham, R. Isolation and characterization of toxins from Xenorhabdus nematophilus against Ferrisia virgata (Ckll.) on tuberose, Polianthes tuberosa. Toxicon 2018, 146, 42–49.
- Mahmood, S.; Kumar, M.; Kumari, P.; Mahapatro, G.K.; Banerjee, N.; Sarin, N.B. Novel insecticidal chitinase from the insect pathogen Xenorhabdus nematophila. Int. J. Biol. Macromol. 2020, 159, 394–401.
- da Silva, W.J.; Pilz-Júnior, H.L.; Heermann, R.; da Silva, O.S. The great potential of entomopathogenic bacteria Xenorhabdus and Photorhabdus for mosquito control: A review. Parasit Vectors 2020, 13, 376.
- Alotaibi, S.S.; Darwish, H.; Alharthi, S.; Alghamdi, A.; Noureldeen, A.; Fallatah, A.M.; Fodor, A.; Al-Barty, A.; Albogami, B.; Baazeem, A. Control potentials of three entomopathogenic bacterial isolates for the carob moth, Ectomyelois ceratoniae (Lepidoptera: Pyralidae) in pomegranates. Agriculture 2021, 11, 1256.
- Bi, Y.; Gao, C.; Yu, Z. Rhabdopeptides from Xenorhabdus budapestensis SN84 and their nematicidal activities against Meloidogyne incognita. J. Agric. Food Chem. 2018, 66, 3833–3839.
- Thaler, J.O.; Baghdiguian, S.; Boemare, N. Purification and characterization of xenorhabdicin, a phage tail-like bacteriocin, from the lysogenic strain F1 of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 1995, 61, 2049–2052.
- Fodor, A.; Fodor, A.M.; Forst, S.; Hogan, J.S.; Klein, M.G.; Lengyel, K.; Sáringer, G.; Stackebrandt, E.; Taylor, R.A.J.; Lehoczky, E. Comparative analysis of antibacterial activities of Xenorhabdus species on related and nonrelated bacteria in vivo. J. Microbiol. Antimicrob. 2010, 2, 36–46.
- Tóth, E.M.; Márialigeti, K.; Fodor, A.; Lucskai, A.; Farkas, R. Evaluation of efficacy of entomopathogenic nematodes against larvae of Lucilia sericata (Meigen, 1826) (Diptera: Calliphoridae). Acta Vet. Hung. 2005, 53, 65–71.
- Furgani, G.; Böszörményi, E.; Fodor, A.; Máthé-Fodor, A.; Forst, S.; Hogan, J.S.; Katona, Z.; Klein, M.G.; Stackebrandt, E.; Szentirmai, A.; et al. Xenorhabdus antibiotics: A comparative analysis and potential utility for controlling mastitis caused by bacteria. J. Appl. Microbiol. 2008, 104, 745–758.
- Thappeta, K.R.V.; Ciezki, K.; Morales-Soto, N.; Wesener, S.; Goodrich-Blair, H.; Stock, S.P.; Forst, S. R-type bacteriocins of Xenorhabdus bovienii determine the outcome of interspecies competition in a natural host environment. Microbiology 2020, 166, 1074–1087.
- Watzel, J.; Hacker, C.; Duchardt-Ferner, E.; Bode, H.B.; Wöhnert, J. A new docking domain type in the peptide-antimicrobial-Xenorhabdus peptide producing nonribosomal peptide synthetase from Xenorhabdus bovienii. ACS Chem. Biol. 2020, 15, 982–989.
- Gualtieri, M.; Aumelas, A.; Thaler, J.O. Identification of a new antimicrobial lysine-rich cyclolipopeptide family from Xenorhabdus nematophila. J. Antibiot. 2009, 62, 295–302.
- Tobias, N.J.; Heinrich, A.K.; Eresmann, H.; Wright, P.R.; Neubacher, N.; Backofen, R.; Bode, H.B. Photorhabdus-nematode symbiosis is dependent on hfq-mediated regulation of secondary metabolites. Environ. Microbiol. 2017, 19, 119–129.
- Bode, E.; Heinrich, A.K.; Hirschmann, M.; Abebew, D.; Shi, Y.N.; Vo, T.D.; Wesche, F.; Shi, Y.M.; Grün, P.; Simonyi, S.; et al. Promoter activation in Δhfq mutants as an efficient tool for specialized metabolite production enabling direct bioactivity testing. Angew. Chem. Int. Ed. Engl. 2019, 58, 18957–18963.
- McErlean, M.; Overbay, J.; Van Lanen, S. Refining and expanding nonribosomal peptide synthetase function and mechanism. J. Ind. Microbiol. Biotechnol. 2019, 46, 493–513.
- Watzel, J.; Sarawi, S.; Duchardt-Ferner, E.; Bode, H.B.; Wöhnert, J. NMR resonance assignments for a docking domain pair with an attached thiolation domain from the PAX peptide-producing NRPS from Xenorhabdus cabanillasii. Biomol. NMR Assign. 2021, 15, 229–234.
- Fuchs, S.W.; Proschak, A.; Jaskolla, T.W.; Karas, M.; Bode, H.B. Structure elucidation and biosynthesis of lysine-rich cyclic peptides in Xenorhabdus nematophila. Org. Biomol. Chem. 2011, 9, 3130–3132.
- Fuchs, S.W.; Sachs, C.C.; Kegler, C.; Nollmann, F.I.; Karas, M.; Bode, H.B. Neutral loss fragmentation pattern based screening for arginine-rich natural products in Xenorhabdus and Photorhabdus. Anal. Chem. 2012, 84, 6948–6955.
- Fuchs, S.W.; Grundmann, F.; Kurz, M.; Kaiser, M.; Bode, H.B. Fabclavines: Bioactive peptide-polyketide-polyamino hybrids from Xenorhabdus. Chembiochem 2014, 15, 512–516.
- Fodor, E.; Szállás, E.; Kiss, Z.; Fodor, A.; Horvath, L.I.; Chitwood, D.J.; Farkas, T. Composition and biophysical properties of lipids in Xenorhabdus nematophilus and Photorhabdus luminescens, symbiotic bacteria associated with entomopathogenic nematodes. Appl. Environ. Microbiol. 1997, 3, 2826–2831.
- Gualtieri, M.; Villain-Guillot, P.; Givaudan, A.; Pages, S. Nemaucin, an Antibiotic Produced by Entomopathogenic Xenorhabdus cabanillasii. France Patent WO2012085177A1, 28 June 2012.
- Wenski, S.L.; Kolbert, D.; Grammbitter, G.L.C.; Bode, H.B. Fabclavine biosynthesis in X. szentirmaii: Shortened derivatives and characterization of the thioester reductase FclG and the condensation domain-like protein FclL. J. Ind. Microbiol. Biotechnol. 2019, 46, 565–572.
- Wenski, S.L.; Cimen, H.; Berghaus, N.; Fuchs, S.W.; Hazir, S.; Bode, H.B. Fabclavine diversity in Xenorhabdus bacteria. Beilstein J. Org. Chem. 2020, 16, 956–965.
- Mollah, M.M.I.; Roy, M.C.; Choi, D.Y.; Hasan, M.A.; Al Baki, M.A.; Yeom, H.S.; Kim, Y. Variations of indole metabolites and NRPS-PKS loci in two different virulent strains of Xenorhabdus hominickii. Front. Microbiol. 2020, 11, 583594.
- Cowles, K.N.; Cowles, C.E.; Richards, G.R.; Martens, E.C.; Goodrich-Blair, H. The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cell Microbiol. 2007, 9, 1311–1323.
- Cao, M.; Patel, T.; Rickman, T.; Goodrich-Blair, H.; Hussa, E.A. High levels of the Xenorhabdus nematophila transcription factor Lrp promote mutualism with the Steinernema carpocapsae nematode host. Appl. Environ. Microbiol. 2017, 83, e00276-17.
- Engel, Y.; Windhorst, C.; Lu, X.; Goodrich-Blair, H.; Bode, H.B. The global regulators Lrp, LeuO, and HexA control secondary metabolism in entomopathogenic bacteria. Front. Microbiol. 2017, 8, 209.
- Ziegler, C.A.; Freddolino, P.L. The leucine-responsive regulatory proteins/ feast-famine regulatory proteins: An ancient and complex class of transcriptional regulators in bacteria and archaea. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 373–400.
- Bhat, A.H.; Chaubey, A.K.; Půža, V. The first report of Xenorhabdus indica from Steinernema pakistanense: Co-phylogenetic study suggests co-speciation between X. indica and its steinernematid nematodes. J. Helminthol. 2019, 93, 81–90.
- Fodor, A.; Forst, S.; Haynes, L.; Hevesi, M.; Hogan, J.A.; Klein, M.G.; Máthe-Fodor, A.; Stackebrandt, E.; Szentirmai, A.; Sztaricskai, F.; et al. New Perspectives of Xenorhabdus Antibiotics Research; Insect Pathogens and Insect Parasitic Nematodes IOBC/IOBC/WPRS Bulletin: Ales, France, 2008; Volume 31, pp. 157–164.
- Hurlbert, R.E.; Xu, J.; Small, C.L. Colonial and cellular polymorphism in Xenorhabdus luminescens. Appl Environ Microbiol. 1989, 55, 1136–1143.
- Givaudan, A.; Baghdiguian, S.; Lanois, A.; Boemare, N. Swarming and swimming changes concomitant with phase variation in Xenorhabdus nematophilus. Appl. Environ. Microbiol. 1995, 1, 1408–1413.
- Givaudan, A.; Lanois, A. FlhDC, the flagellar master operon of Xenorhabdus nematophilus: Requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J. Bacteriol. 2000, 182, 107–115.
- Kim, D.J.; Boylan, B.; George, N.; Forst, S. Inactivation of ompR promotes precocious swarming and FlhDC expression in Xenorhabdus nematophila. J. Bacteriol. 2003, 185, 5290–5294.
- Park, D.; Forst, S. Co-regulation of motility, exoenzyme and antibiotic production by the EnvZ-OmpR-FlhDC-FliA pathway in Xenorhabdus nematophila. Mol. Microbiol. 2006, 61, 1397–1412.
- Chandra, H.; Khandelwal, P.; Khattri, A.; Banerjee, N. Type 1 fimbriae of insecticidal bacterium Xenorhabdus nematophila is necessary for growth and colonization of its symbiotic host nematode Steinernema carpocapsiae. Environ. Microbiol. 2008, 10, 1285–1295.
- Fodor, A.; Varga, I.; Hevesi, M.; Máthé-Fodor, A.; Racsko, J.; Hogan, J.A. Novel anti-microbial peptides of Xenorhabdus origin against multidrug resistant plant pathogens. In A Search for Antibacterial Agents; Bobbarala, V., Ed.; IntechOpen: London, UK, 2012; pp. 3–32. Available online: https://www.intechopen.com/books/2129 (accessed on 28 February 2022).
- Hanson, A.W.; Hum, L.K. The crystal structure of. iodinin. Acta Cryst. 1969, 25, 768.
- Myhren, L.E.; Nygaard, G.; Gausdal, G.; Sletta, H.; Teigen, K.; Degnes, K.F.; Zahlsen, K.; Brunsvik, A.; Bruserud, Ø.; Døskeland, S.O.; et al. Iodinin (1,6-dihydroxyphenazine 5,10-dioxide) from Streptosporangium sp. induces apoptosis selectively in myeloid leukemia cell lines and patient cells. Mar. Drugs 2013, 11, 332–349.
- Sletta, H.; Degnes, K.F.; Herfindal, L.; Klinkenberg, G.; Fjærvik, E.; Zahlsen, K.; Brunsvik, A.; Nygaard, G.; Aachmann, F.L.; Ellingsen, T.E.; et al. Anti-microbial and cytotoxic 1,6-dihydroxyphenazine-5,10-dioxide (iodinin) produced by Streptosporangium sp. DSM 45942 isolated from the fjord sediment. Appl. Microbiol. Biotechnol. 2014, 98, 603–610.
- Viktorsson, E.Ö.; Melling Grøthe, B.; Aesoy, R.; Sabir, M.; Snellingen, S.; Prandina, A.; Høgmoen Åstrand, O.A.; Bonge-Hansen, T.; Døskeland, S.O.; Herfindal, L.; et al. Total synthesis and antileukemic evaluations of the phenazine 5,10-dioxide natural products iodinin, myxin and their derivatives. Bioorg. Med. Chem. 2017, 25, 2285–2293.
- Viktorsson, E.Ö.; Aesoy, R.; Støa, S.; Lekve, V.; Døskeland, S.O.; Herfindal, L.; Rongved, P. New prodrugs and analogs of the phenazine 5,10-dioxide natural products iodinin and myxin promote selective cytotoxicity towards human acute myeloid leukemia cells. RSC Med. Chem. 2021, 12, 767–778.
- Prandina, A.; Herfindal, L.; Radix, S.; Rongved, P.; Døskeland, S.O.; Le Borgne, M.; Perret, F. Enhancement of iodinin solubility by encapsulation into cyclodextrin nanoparticles. J. Enzyme Inhib. Med. Chem. 2018, 33, 370–375.
- SMART 5.630; Bruker Advanced X-ray Solutions. Bruker AXS Inc.: Madison, WI, USA, 2005.
- SAINT V8.40B; Bruker Advanced X-ray Solutions. Bruker AXS Inc.: Madison, WI, USA, 2020.
- Krause, L.; Herbst-Irmer, R.; Sheldrick, G.M.; Stalke, D. Comparison of silver and molybdenum microfocus X-ray sources for single-crystal structure determination. J. Appl. Crystallogr. 2015, 48 Pt 1, 3–10.
- Hübschle, C.B.; Sheldrick, G.M.; Dittrich, B. ShelXle: A Qt graphical user interface for SHELXL. J. Appl. Crystallogr. 2011, 44 Pt 6, 1281–1284.
- SHELXTL Suite of Programs; Version 6.14, 2000–2003; Bruker Advanced X-ray Solutions; Bruker AXS Inc.: Madison, WI, USA, 2008.
- Sheldrick, G.M. A Short history of SHELX. Acta Crystallogr. 2008, 64, 112–122.
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2018, 71, 3–8.
- Lübben, J.; Wandtke, C.M.; Hübschle, C.B.; Ruf, M.; Sheldrick, G.M.; Dittrich, B. Aspherical scattering factors for SHELXL - model, implementation and application. Acta. Crystallogr. A Found Adv. 2019, 75 Pt 1, 50–62.
- Pang, B.; Liu, T.; Zhang, W.; Ye, F.; Shang, C. Cloning and characterization of phzR gene from Pseudomonas aeruginosa. Curr. Microbiol. 2021, 78, 1482–1487.
- Shi, Y.M.; Brachmann, A.O.; Westphalen, M.A.; Neubacher, N.; Tobias, N.J.; Bode, H.B. Dual phenazine gene clusters enable diversification during biosynthesis. Nat. Chem. Biol. 2019, 15, 331–339.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
3 times
(View History)
Update Date:
20 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




