
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Bingyun Sun | -- | 2548 | 2022-04-07 22:08:06 | | | |
| 2 | Jason Zhu | -62 word(s) | 2486 | 2022-04-08 04:48:51 | | |
Video Upload Options
N-Glycosylation (NG) and disulfide bonds (DBs) are two prevalent co/post-translational modifications (PTMs) that are often conserved and coexist in membrane and secreted proteins involved in a large number of diseases. Both in the past and in recent times, the enzymes and chaperones regulating these PTMs have been constantly discovered to directly interact with each other or colocalize in the ER. However, beyond a few model proteins, how such cooperation affects N-glycan modification and disulfide bonding at selective sites in individual proteins is largely unknown. More investigations should be encouraged to unveil the hidden relationships of NG and DBs in the majority of membranes and secreted proteins for pathophysiological understanding and biotherapeutic development.
1. Introduction
2. Promoting Relationship
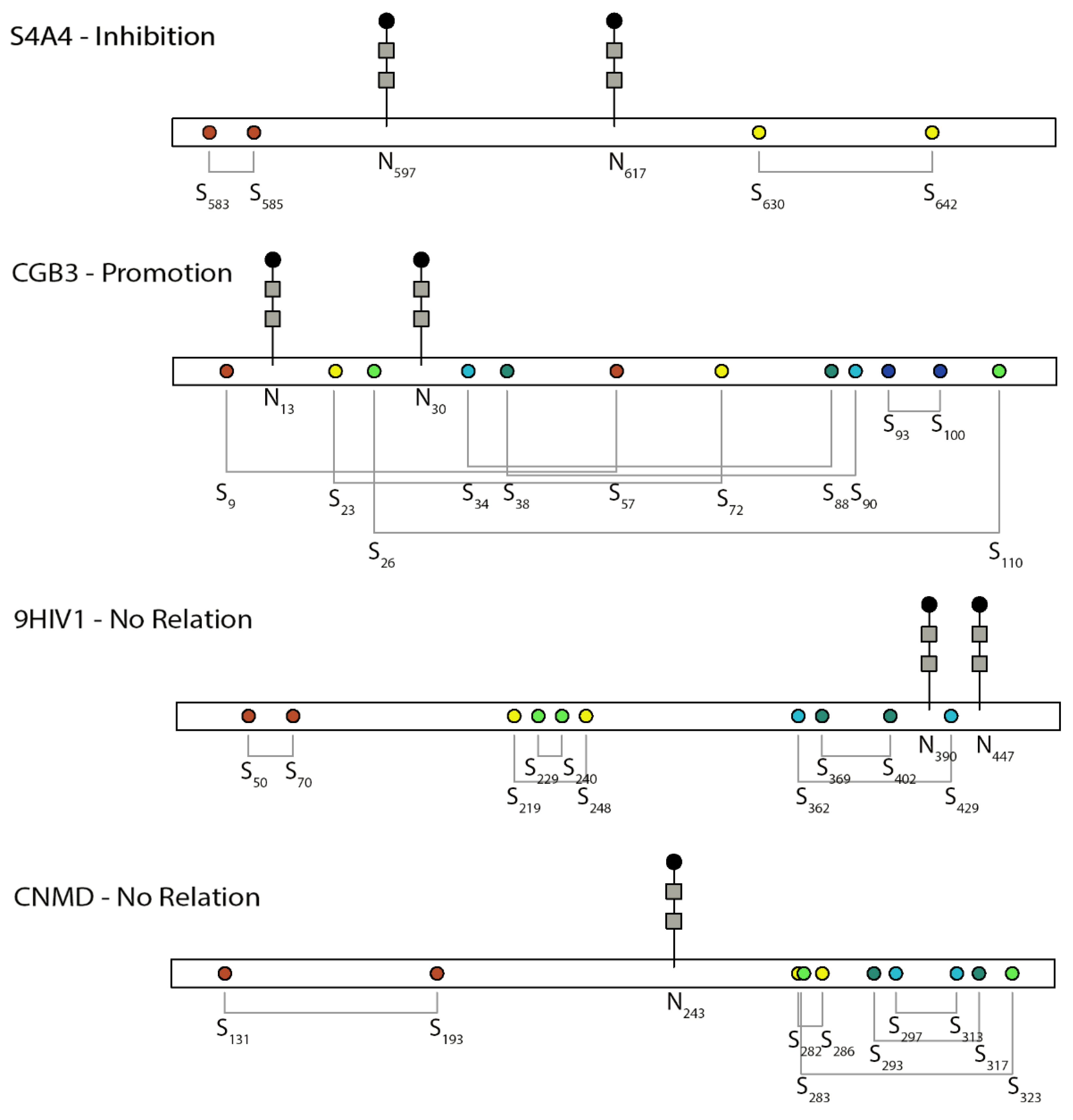
3. Inhibitory Relationship
4. Independent Relationship
5. Unknown Relation
References
- Adams, B.M.; Oster, M.E.; Hebert, D.N. Protein Quality Control in the Endoplasmic Reticulum. Protein J. 2019, 38, 317–329.
- Grek, C.; Townsend, D.M. Protein Disulfide Isomerase Superfamily in Disease and the Regulation of Apoptosis. Endoplasmic Reticulum Stress Dis. 2014, 1, 4–17.
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366.
- Feng, W.; Huth, J.R.; Norton, S.E.; Ruddon, R.W. Asparagine-linked oligosaccharides facilitate human chorionic gonadotropin beta-subunit folding but not assembly of prefolded beta with alpha. Endocrinology 1995, 136, 52–61.
- Moriwaki, T.; Suganuma, N.; Furuhashi, M.; Kikkawa, F.; Tomoda, Y.; Boime, I.; Nakata, M.; Mizuochi, T. Alteration of N-linked oligosaccharide structures of human chorionic gonadotropin beta-subunit by disruption of disulfide bonds. Glycoconj. J. 1997, 14, 225–229.
- Beggah, A.T.; Jaunin, P.; Geering, K. Role of glycosylation and disulfide bond formation in the beta subunit in the folding and functional expression of Na, K-ATPase. J. Biol. Chem. 1997, 272, 10318–10326.
- Laughery, M.D.; Todd, M.L.; Kaplan, J.H. Mutational analysis of alpha-beta subunit interactions in the delivery of Na, K-ATPase heterodimers to the plasma membrane. J. Biol. Chem. 2003, 278, 34794–34803.
- Mirazimi, A.; Svensson, L. Carbohydrates facilitate correct disulfide bond formation and folding of rotavirus VP7. J. Virol. 1998, 72, 3887–3892.
- Asker, N.; Axelsson, M.A.; Olofsson, S.O.; Hansson, G.C. Dimerization of the human MUC2 mucin in the endoplasmic reticulum is followed by a N-glycosylation-dependent transfer of the mono- and dimers to the Golgi apparatus. J. Biol. Chem. 1998, 273, 18857–18863.
- McKinnon, T.A.; Goode, E.C.; Birdsey, G.M.; Nowak, A.A.; Chan, A.C.; Lane, D.A.; Laffan, M.A. Specific N-linked glycosylation sites modulate synthesis and secretion of von Willebrand factor. Blood 2010, 116, 640–648.
- Ishmael, S.S.; Ishmael, F.T.; Jones, A.D.; Bond, J.S. Protease domain glycans affect oligomerization, disulfide bond formation, and stability of the meprin A metalloprotease homo-oligomer. J. Biol. Chem. 2006, 281, 37404–37415.
- Daniels, R.; Kurowski, B.; Johnson, A.E.; Hebert, D.N. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol. Cell 2003, 11, 79–90.
- UniProt Consortium, T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699.
- McGinnes, L.W.; Morrison, T.G. Disulfide bond formation is a determinant of glycosylation site usage in the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus. J. Virol. 1997, 71, 3083–3089.
- McGinnes, L.W.; Wilde, A.; Morrison, T.G. Nucleotide sequence of the gene encoding the Newcastle disease virus hemagglutinin-neuraminidase protein and comparisons of paramyxovirus hemagglutinin-neuraminidase protein sequences. Virus Res. 1987, 7, 187–202.
- McGinnes, L.W.; Morrison, T.G. The role of the individual cysteine residues in the formation of the mature, antigenic HN protein of Newcastle disease virus. Virology 1994, 200, 470–483.
- Mirza, A.M.; Sheehan, J.P.; Hardy, L.W.; Glickman, R.L.; Iorio, R.M. Structure and function of a membrane anchor-less form of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus. J. Biol. Chem. 1993, 268, 21425–21431.
- Allen, S.; Naim, H.Y.; Bulleid, N.J. Intracellular folding of tissue-type plasminogen activator. Effects of disulfide bond formation on N-linked glycosylation and secretion. J. Biol. Chem. 1995, 270, 4797–4804.
- Kao, L.; Sassani, P.; Azimov, R.; Pushkin, A.; Abuladze, N.; Peti-Peterdi, J.; Liu, W.; Newman, D.; Kurtz, I. Oligomeric structure and minimal functional unit of the electrogenic sodium bicarbonate cotransporter NBCe1-A. J. Biol. Chem. 2008, 283, 26782–26794.
- Zhu, Q.; Kao, L.; Azimov, R.; Abuladze, N.; Newman, D.; Kurtz, I. Interplay between disulfide bonding and N-glycosylation defines SLC4 Na+-coupled transporter extracellular topography. J. Biol. Chem. 2015, 290, 5391–5404.
- Bolmstedt, A.; Hemming, A.; Flodby, P.; Berntsson, P.; Travis, B.; Lin, J.P.; Ledbetter, J.; Tsu, T.; Wigzell, H.; Hu, S.L.; et al. Effects of mutations in glycosylation sites and disulphide bonds on processing, CD4-binding and fusion activity of human immunodeficiency virus envelope glycoproteins. J. Gen. Virol. 1991, 72 Pt 6, 1269–1277.
- Hemming, A.; Bolmstedt, A.; Flodby, P.; Lundberg, L.; Gidlund, M.; Wigzell, H.; Olofsson, S.O. Cysteine 402 of HIV gp120 is essential for CD4-binding and resistance of gp120 to intracellular degradation. Arch. Virol. 1989, 109, 269–276.
- Tschachler, E.; Buchow, H.; Gallo, R.C.; Reitz, M.S., Jr. Functional contribution of cysteine residues to the human immunodeficiency virus type 1 envelope. J. Virol. 1990, 64, 2250–2259.
- Kondo, J.; Shibata, H.; Miura, S.; Yamakawa, A.; Sato, K.; Higuchi, Y.; Shukunami, C.; Hiraki, Y. A functional role of the glycosylated N-terminal domain of chondromodulin-I. J. Bone Miner. Metab. 2011, 29, 23–30.
- Norskov-Lauritsen, L.; Jorgensen, S.; Brauner-Osborne, H. N-glycosylation and disulfide bonding affects GPRC6A receptor expression, function, and dimerization. FEBS Lett. 2015, 589, 588–597.
- Pi, M.; Nishimoto, S.K.; Quarles, L.D. GPRC6A: Jack of all metabolism (or master of none). Mol. Metab. 2017, 6, 185–193.
- Clemmensen, C.; Smajilovic, S.; Wellendorph, P.; Brauner-Osborne, H. The GPCR, class C, group 6, subtype A (GPRC6A) receptor: From cloning to physiological function. Br. J. Pharmacol. 2014, 171, 1129–1141.
- Muto, T.; Tsuchiya, D.; Morikawa, K.; Jingami, H. Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc. Natl. Acad. Sci. USA 2007, 104, 3759–3764.
- Rider, P.J.F.; Naderi, M.; Bergeron, S.; Chouljenko, V.N.; Brylinski, M.; Kousoulas, K.G. Cysteines and N-Glycosylation Sites Conserved among All Alphaherpesviruses Regulate Membrane Fusion in Herpes Simplex Virus 1 Infection. J. Virol. 2017, 91, e00873-17.
- Neubauer, A.; Osterrieder, N. Equine herpesvirus type 1 (EHV-1) glycoprotein K is required for efficient cell-to-cell spread and virus egress. Virology 2004, 329, 18–32.
- Zhang, S.; Go, E.P.; Ding, H.; Anang, S.; Kappes, J.C.; Desaire, H.; Sodroski, J. Analysis of glycosylation and disulfide bonding of wild-type SARS-CoV-2 spike glycoprotein. J. Virol. 2022, 96, e01626-21.
- Brun, J.; Vasiljevic, S.; Gangadharan, B.; Hensen, M.; Chandran, A.V.; Hill, M.L.; Kiappes, J.L.; Dwek, R.A.; Alonzi, D.S.; Struwe, W.B.; et al. Analysis of SARS-CoV-2 spike glycosylation reveals shedding of a vaccine candidate. bioRxiv 2020.
- Wang, D.; Baudys, J.; Bundy, J.L.; Solano, M.; Keppel, T.; Barr, J.R. Comprehensive Analysis of the Glycan Complement of SARS-CoV-2 Spike Proteins Using Signature Ions-Triggered Electron-Transfer/Higher-Energy Collisional Dissociation (EThcD) Mass Spectrometry. Anal. Chem. 2020, 92, 14730–14739.
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738.e13.
- Sanda, M.; Morrison, L.; Goldman, R. N and O glycosylation of the SARS-CoV-2 spike protein. Anal. Chem. 2021, 93, 2003–2009.
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333.
- Bangaru, S.; Ozorowski, G.; Turner, H.L.; Antanasijevic, A.; Huang, D.; Wang, X.; Torres, J.L.; Diedrich, J.K.; Tian, J.H.; Portnoff, A.D.; et al. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science 2020, 370, 1089–1094.
- Zhao, P.; Praissman, J.L.; Grant, O.C.; Cai, Y.; Xiao, T.; Rosenbalm, K.E.; Aoki, K.; Kellman, B.P.; Bridger, R.; Barouch, D.H.; et al. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell Host Microbe 2020, 28, 586–601 e586.
- Hati, S.; Bhattacharyya, S. Impact of Thiol-Disulfide Balance on the Binding of Covid-19 Spike Protein with Angiotensin-Converting Enzyme 2 Receptor. ACS Omega 2020, 5, 16292–16298.
- Singh, J.; Dhindsa, R.S.; Misra, V.; Singh, B. SARS-CoV2 infectivity is potentially modulated by host redox status. Comput. Struct. Biotechnol. J. 2020, 18, 3705–3711.
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220.
- Cherepanova, N.A.; Shrimal, S.; Gilmore, R. Oxidoreductase activity is necessary for N-glycosylation of cysteine-proximal acceptor sites in glycoproteins. J. Cell Biol. 2014, 206, 525–539.
- Mehdipour, A.R.; Hummer, G. Dual nature of human ACE2 glycosylation in binding to SARS-CoV-2 spike. Proc. Natl. Acad. Sci. USA 2021, 118, e2100425118.
- Vlahos, C.J.; Wilhelm, O.G.; Hassell, T.; Jaskunas, S.R.; Bang, N.U. Disulfide pairing of the recombinant kringle-2 domain of tissue plasminogen activator produced in Escherichia coli. J. Biol. Chem. 1991, 266, 10070–10072.
- Marti, T.; Rosselet, S.J.; Titani, K.; Walsh, K.A. Identification of disulfide-bridged substructures within human von Willebrand factor. Biochemistry 1987, 26, 8099–8109.
- Liu, T.; Qian, W.J.; Gritsenko, M.A.; Camp, D.G., 2nd; Monroe, M.E.; Moore, R.J.; Smith, R.D. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 2005, 4, 2070–2080.




