Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Arthur BAGEL | -- | 3561 | 2022-04-05 09:53:31 | | | |
| 2 | Conner Chen | -16 word(s) | 3545 | 2022-04-06 03:36:16 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Bagel, A.; Sergentet, D. Shiga Toxin-Producing Escherichia coli and Milk Fat Globules. Encyclopedia. Available online: https://encyclopedia.pub/entry/21363 (accessed on 07 February 2026).
Bagel A, Sergentet D. Shiga Toxin-Producing Escherichia coli and Milk Fat Globules. Encyclopedia. Available at: https://encyclopedia.pub/entry/21363. Accessed February 07, 2026.
Bagel, Arthur, Delphine Sergentet. "Shiga Toxin-Producing Escherichia coli and Milk Fat Globules" Encyclopedia, https://encyclopedia.pub/entry/21363 (accessed February 07, 2026).
Bagel, A., & Sergentet, D. (2022, April 05). Shiga Toxin-Producing Escherichia coli and Milk Fat Globules. In Encyclopedia. https://encyclopedia.pub/entry/21363
Bagel, Arthur and Delphine Sergentet. "Shiga Toxin-Producing Escherichia coli and Milk Fat Globules." Encyclopedia. Web. 05 April, 2022.
Copy Citation
Shiga toxin-producing Escherichia coli (STEC) are zoonotic Gram-negative bacteria. While raw milk cheese consumption is healthful, contamination with pathogens such as STEC can occur due to poor hygiene practices at the farm level. STEC infections cause mild to serious symptoms in humans. The raw milk cheese-making process concentrates certain milk macromolecules such as proteins and milk fat globules (MFGs), allowing the intrinsic beneficial and pathogenic microflora to continue to thrive. MFGs are surrounded by a biological membrane, the milk fat globule membrane (MFGM), which has a globally positive health effect, including inhibition of pathogen adhesion.
STEC
MFGM
Milk Fat Globules
Raw milk Cheese
bacterial adhesion
1. Introduction
In the Code of Hygienic Practice for Milk and Milk Products (Codex Alimentarius, 2004), raw milk is defined as milk that has not been heated beyond 40 °C or undergone any treatment that has an equivalent effect. Raw milk is an oil-in-water type emulsion and contains almost 900 g of water and 130 g of dry matter, in varying proportions [1]. Milk is a major source of calcium, and also an important supply of proteins, for those who consume it (newborn mammals and humans). Milk contains all essential amino acids, lipids, fatty acids, vitamins, and lactose [2]. One liter of whole milk contains approximately 38 g of fat, which is present mainly in the form of milk fat globules (MFGs) [3].
Raw milk cheeses are made from raw cow, sheep, or goat milk. Different cheese-making processes can be applied to create the end-products. The various combinations of ingredients (such as milk and cultures) and cheese-making processes result in a large diversity of cheeses. No less than 1200 different cheeses are made in France [4]. These include ripened or unripened soft, semi-hard, hard, or extra-hard products, which may be coated, uncooked, or cooked pressed cheeses (with short or long ripening), blue-type cheeses, lactic cheeses, and white mold cheeses.
Raw milk is unstable due to the presence of microflora and, therefore, is often treated to optimize its conservation and to prevent microbiological hazards. The microbiota of raw milk is complex and is derived from many sources, including direct contact with the animal (teats, hides, and feces), the surface of milking equipment (particularly if hygiene practices are poor), housing, bedding, feed, air, and water [5][6][7]. On the one hand, this dynamic bacterial community plays several beneficial roles in subsequent dairy products such as facilitating fermentation and promoting the health of consumers. On the other hand, microbiota can contain spoilage or pathogenic bacteria [5]. The milk microbiota is almost eliminated during heat treatment, such as ultra-high temperature (UHT) sterilization, and microfiltration, while in raw milk the microbiota is preserved. Different studies have shown that the raw milk microbiota is useful to the human digestive system; therefore, it may be beneficial to preserve it [5][8]. In addition, the raw milk microbiota gives raw milk cheeses more varied and intense flavors than heat-treated milk cheeses. The raw milk microbiota can also influence the human intestinal microbiota, which contributes significantly to human health, for example, by modulating the immune system. The consumption of raw milk and raw milk cheeses can also reduce blood pressure in people with mild to moderate hypertension [9] and decrease allergies in infants [10][11]. However, the risk–benefit ratio of consuming raw milk products is difficult to estimate. Nevertheless, in France, it is strongly recommended to avoid giving raw milk products to children under 5 years old, pregnant women, and immunocompromised patients [12]. If poor hygiene procedures have been applied, particularly during milking, raw milk may be contaminated by pathogenic bacteria such as Shiga toxin-producing Escherichia coli (STEC).
STEC are foodborne zoonotic bacteria associated with large-scale epidemics that represent a major public health problem. Human STEC infection is most often linked to the ingestion of contaminated food and water, such as undercooked ground meat, raw milk cheeses, or raw vegetables. Raw milk and raw milk cheeses have been linked to foodborne infections associated with STEC in humans from different countries [13][14][15][16][17][18][19][20]. Ruminants are the primary reservoir of STEC. Milk is most often contaminated by feces (directly or indirectly) during the milking process. STEC are very frequently associated with severe forms of infection such as hemorrhagic colitis and, in very severe cases, systemic complications including hemolytic uremic syndrome (HUS). HUS is the leading cause of renal failure in children under 3 years of age. The estimated infectious dose is very low: between 5 and 50 viable cells [21][22].
The proportion of milk and dairy products involved in Rapid Alert System for Food and Feed (RASFF) notifications issued due to food contamination with STEC is very low compared to those involving meat products [23]. The RASFF is a European communication tool used when public health microbiological hazards are detected in the food chain and food products. In 2013, two RASFF notifications were related to STEC-contaminated dairy products versus 68 for meat products. In 2014, four RASFF notifications of dairy products contaminated with STEC were listed, compared with 53 for meat products. Likewise, there were 7 versus 16, 8 versus 26, and 4 versus 49 dairy-related and meat-related notifications, respectively, in 2015, 2016, and 2017 [23]. Furthermore, epidemiological studies have shown that this class of product is only a minor source of human enteric infection [24][25][26]. Interestingly, prevalence data on these enteropathogens in dairy matrices and ingestion-related outbreaks do not fit overall foodborne-related outbreak figures [26]. A study led by Douëllou et al. [27] showed that there were no differences in the key virulence properties of dairy STEC isolates compared with human isolates. The same authors hypothesized that this phenomenon might be related to an association between STEC and MFGs, thus inhibiting STEC adhesion to enterocytes.
Milk fat globules (MFGs) have a positive impact on the immune system, and their antimicrobial properties have been largely described [28][29][30][31]. The positive action of MFGs on human health seems to be carried out by the membrane (and membrane components) surrounding the globules. MFGs are small lipid droplets formed by a core of triacylglycerols (TAGs) and enveloped by a biological phospholipid triple membrane, the milk fat globule membrane (MFGM), which is derived from mammary epithelial cells [32]. The outer bilayer of the MFGM contains diverse (glyco)-proteins and (glyco)-lipids on its surface [33]. These glycoconjugates make up the glycocalyx and act as a source of specific bacterial and viral ligands [29][34].
2. Raw Milk Sector and STEC
2.1. Importance of Raw Milk Cheeses
Cheeses are products with high added value and significant economic importance in France and Europe. In 2019, the annual cheese consumption per inhabitant was 26.8 kg in France and 19.1 kg in all of Europe [35]. In total, 10,630,000 t of cheese were produced in Europe in 2019 [35]. In France, cheeses represent one of the main food industries, worth approximately EUR 38.7 billion in 2017 [36]. In 2019, with all milk processing combined, French cheese production included 1,664,632 t of cow’s milk cheese, 99,265 t of goat’s milk cheese, and 59,638 t of sheep’s milk cheese [35]. The production of raw milk cheese accounted for 172,128 t of the cow’s milk cheese, 20,872 t of the sheep’s milk cheese, and 9691 t of the goat’s milk cheese produced in France in 2019 [35]. Raw milk cheeses represent the vast majority of farmhouse dairy products and approximately 75% of the volume of cheeses marketed under quality and origin identification signs (SIQO), including protected designations of origin (PDO) and protected geographical indications (PGI). Finally, a study has shown that 75% of French people consume raw milk cheeses at least once a month and that 33% of French people consume raw milk cheeses every week [37]. In contrast, no data on the worldwide consumption or production of raw milk cheeses are currently available.
Raw milk cheeses are part of French and European food heritage and are an essential tool to enhance product value and to create dynamism in our territories. They are part of a dynamic of rural development and land use planning; their production avoids the desertification of certain areas by providing a significant source of income for farmers. They are often produced under a PDO quality label. Raw milk cheeses encourage variety and diversification of production and contribute to the sustainability of rural economies. They protect traditional production areas, enhance the recognized know-how of operators, and facilitate, especially for small producers, the marketing of differentiated products with specific and clearly identifiable characteristics. In France, the competent authorities as well as scientists recognize the importance of raw milk cheese both in terms of gastronomic heritage and regional socio-economic development. At the same time, authorities and scientists seek to support the industry by improving scientific knowledge about STEC and developing methods for the control and surveillance of this bacteria from farm to fork.
2.2. STEC
STEC are foodborne zoonotic bacteria associated with large-scale epidemics that represent a major public health problem. Ruminants (cattle, buffalo, goats, and sheep) are the main reservoir of STEC [38][39]. Infected ruminants can be asymptomatic, harboring the bacteria in their gastrointestinal tract and shedding bacteria in their feces [40][41][42]. Detailed investigations have shown that without proper cleaning methods and udder hygiene practices, feces can contaminate the teats and udders of animals and cause milk contamination during milking [43]. When STEC-contaminated milk is used to produce raw milk cheeses, STEC can survive and be isolated in some cheeses.
The pathogenesis of STEC-related disease generally involves three phases: (i) ingestion of contaminated food; (ii) colonization of the intestinal epithelium by STEC; and (iii) production of Shiga toxins (Stx) that disrupt normal cellular functions and damage cells. Stx are the main virulence factors of STEC. The Stx family includes all toxins with a similar structure and biological activity. Based on their different in vitro and in vivo toxicity, amino acid sequences, or nucleotide sequences of the stx genes, two major types of Shiga toxins, Stx1 and Stx2, and numerous variants (Stx1a to Stx1d and Stx2a to Stx2k) have been identified [44][45][46][47]. To effectively colonize a host and cause disease, STEC have evolved mechanisms and strategies for attaching or adhering to host cells and tissues [48]. Adhesion is required so that STEC cells are not swept away by the host’s natural self-cleaning mechanisms. An arsenal of STEC surface adhesion factors have been described (Figure 1).
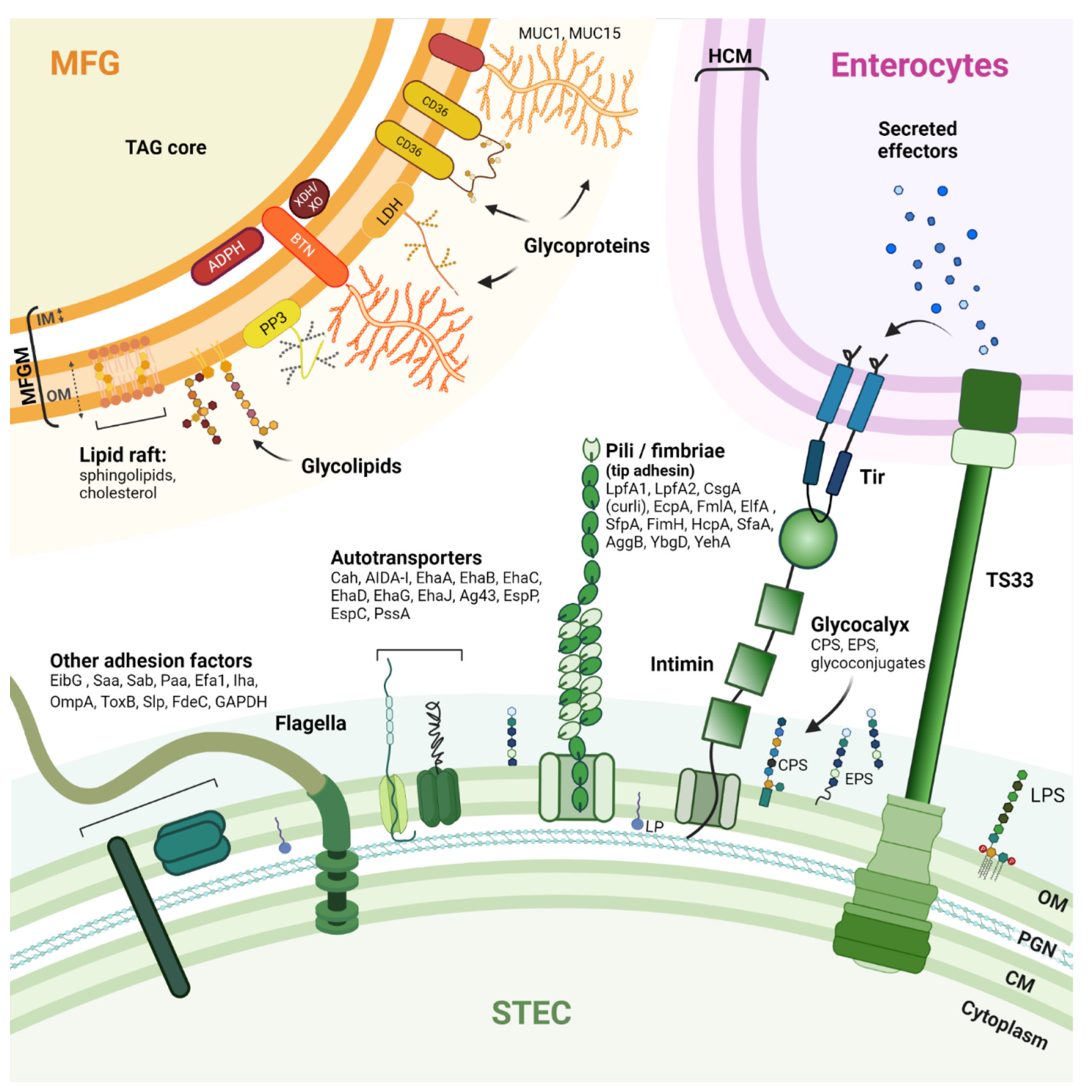
Figure 1. Schematic drawing of STEC adhesion factors and the bovine MFGM. STEC have an arsenal of protein structures involved in host cell adhesion. However, the adhesion mechanisms that contribute to the pathogenicity of STEC are not fully understood, and the receptors recognized by these adhesion factors are not all known. Nevertheless, some pili, autotransporters, and flagella can bind to host extracellular matrix (ECM) components such as fibronectin and laminin (glycoproteins). The MFGM is a complex trilayered structure, comprising a monolayer of polar lipids derived from endoplasmic reticulum (IM: inner membrane) and a bilayer of polar lipids originating from the apical plasma membrane of mammary secretory cells (OM: outer membrane). The structures drawn do not necessarily reflect the actual structures of the macromolecules and are not to scale. CM: Cytoplasmic membrane; PGN: Peptidoglycan; OM: Outer membrane; LPS: Lipopolysaccharide; LP: Lipoprotein; Tir: Translocated intimin receptor; T3SS: Type 3 secretion system; HCM: Host cytoplasmic membrane; CPS: Capsular polysaccharide; EPS: Extracellular polysaccharide; LpfA: Long polar fimbria subunit A; CsgA: Major curlin subunit; EcpA: E. coli common pilus subunit A; FmlA: Type-1 fimbria subunit A; ElfA: laminin-binding fimbria subunit A; SfpA: sorbitol-fermenting fimbria subunit A; FimH: Type 1 fimbrin D-mannose specific adhesin; HcpA: Hemorrhagic coli pilus subunit A; SfaA: S-fimbria subunit A; AggB: Aggregative adherence fimbria I subunit B; YbgD: Putative fimbria Ybg subunit A; YehA: Putative fimbria Yeh subunit A; Cah: Calcium-binding antigen 43 homologue; AIDA-I: Adhesin involved in diffuse adherence; Eha: Enterohaemorrhagic E. coli autotransporter; Ag43: Antigen 43; EspP: Extracellular serine protease plasmid encoded; EspC: EPEC-secreted protein C; PssA: Protease secreted by STEC; EibG: E. coli immunoglobulin-binding protein G; Saa: STEC autoagglutinating adhesion autotransporter; Sab: STEC autotransporter contributing to biofilm formation; Paa: porcine A/E-associated protein; Efa1: EHEC factor for adherence; Iha: IrgA homologue adhesin; OmpA: Outer membrane protein A; ToxB: Toxin B; Slp: Carbon starvation-inducible lipoprotein; FdeC: Factor adherence E. coli; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase. MUC1/MUC15: Mucin 1/15; LDH: Lactadherin; ADPH: Adipophilin; BTN: Butyrophilin; XDH/XO: Xanthine dehydrogenase/oxidase; CD36: Cluster of differentiation 36; PP3: Proteose peptone 3; TAG: triacylglycerols.
The ability to adhere to the intestinal epithelium and colonize the intestine undeniably contributes to the pathogenic processes of STEC cells. Thus, the vast majority of clinical isolates known to cause bloody diarrhea or HUS have one or more virulence factors that allow their adhesion to intestinal epithelial cells [44]. The major adhesion factor of clinical STEC isolates is intimin [49], a protein encoded by the eae gene that resides in the locus of an enterocyte effacement pathogenicity island (LEE). The pathophysiology of clinical isolates possessing the eae gene is characterized by the development of enterocyte attachment-effacement (A/E) lesions. These lesions are responsible for the diarrhea observed in patients [49]. Intimin attachment to the host cell requires an upstream connection of the STEC cell to the host cell cytoplasm [48]. Intimin binds to the translocated intimin receptor (Tir) protein, which is encoded by STEC and translocated into the host cell cytoplasm using a type III secretion system (T3SS) and then inserted into the host cell membrane. Although STEC isolates carrying the eae gene represent the vast majority of human infections, some STEC lacking the eae gene have been isolated from patients [50][51]. An early adhesion phase involving other adhesion factors may occur before, or in parallel with, the formation of the highly specific intimin/Tir bond. Intimin can also bind, with less specificity and strength, to certain host cell surface components such as integrin and nucleolin, and this may contribute to STEC–host cell adhesion [52][53][54]. Some studies suggest involvement of the long polar fimbriae (LPF), which recognizes moieties of eukaryotic extracellular matrix (ECM) components [55][56]. Molecular characterization studies of STEC isolates have also identified paa, efa1, ompA, saa, sab, toxB, and aggR as genes encoding virulence factors involved in adhesion [57][58][59]. Flagella are also involved in STEC adhesion by binding to mucus and mucin proteins [60]. Other proteins can interact with immunoglobulins, for example, E. coli immunoglobulin-binding protein (Eib) [61][62]. The complete list of virulence factors, the timing of their expression, and the mechanisms involved in STEC pathogenicity are not yet fully known. Current knowledge of STEC surface proteins is summarized in Figure 1. STEC adhesion mechanisms are further detailed in these articles: [48][59][63].
2.3. Milk Fat Globules
MFGs can be distinguished from other forms of fat by the milk fat globule membrane (MFGM) that surrounds a core of triacylglycerols (TAGs) (Figure 1). Complex fat supramolecular organizations are also found in egg yolk or oilseeds in the form of oleosomes [64]. The MFGM is made of phospho- and sphingolipids, cholesterol, and proteins [65][66]. As a consequence of the mechanism of milk fat secretion from mammary epithelial cells, the MFGM is a complex trilayered structure, comprising a monolayer of polar lipids derived from the endoplasmic reticulum and a bilayer of polar lipids originating from the apical plasma membrane of the mammary secretory cells [67][68][69]. The MFG size is dependent on the origin of the milk. Bovine MFGs have a mean diameter of 4 µm, while MFGs from goat (3.19 µm), camel (2.99 µm), and sheep raw milk (3.78 µm) are all smaller, and MFGs from buffalo (8.7 µm) are much larger [70]. The main MFGM (glyco)-proteins include glycoproteins mucin 1 and 15 (MUC1; MUC15), the redox enzyme xanthine dehydrogenase/oxidase (XDH/XO), butyrophilin (BTN), cluster of differentiation 36 (CD36), lactadherin (LDH); and two proteins: adipophilin (ADPH) and fatty-acid binding protein (FABP) [29][71].
3. STEC in Raw Milk Cheeses
3.1. Prevalence and Behavior of STEC in Raw Milk Cheeses
The most comprehensive studies on the prevalence of STEC in cheeses have been conducted in Europe and show that the prevalence of STEC varies from 0% to 13.1%, depending on the study [72]. In France in 2009, 2014, and 2018, surveillance plans assessed the prevalence of specific STEC isolates (E. coli possessing the eae and stx genes and belonging to O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28 serotypes) in raw milk cheeses. These studies showed a prevalence of 0.9%, 0.2%, and 0.8% in the raw milk cheese studied in 2009, 2014, and 2018, respectively. In 2016, researchers in a French study evaluated the genetic diversity and virulence gene profiles of STEC isolated from dairy products [27]. They showed that the 197 studied isolates displayed a high genetic diversity regardless of their serotype, with Simpson’s Diversity Index ranging from 1.0 to 0.9615. In addition, their results suggested that the virulence gene profiles of the dairy isolates are a potential hazard. Nevertheless, for the isolate most frequently found in dairy products, O26:H11 STEC, gene expression was similar between human and dairy isolates except for stx1 (44% vs. 87%) and stx2 (81% vs. 23%) expression. It is important to keep in mind that Stx2 has stronger cytotoxicity than Stx1 [73]. A French team showed that during the manufacture of different types of cheese experimentally contaminated with STEC strains, there was no statistically significant strain effect for the same serotype [74][75]. However, only a few different strains were tested for each serotype. Interestingly, a serotype effect was observed in certain types of cheese. Researchers observed less growth of the serotype O157:H7 strains than of the serotype O26:H11, O103:H2, and O145:H28 strains. They hypothesized that strains belonging to serotypes O26:H11, O103:H2, and O145:H28 could be more adapted to the conditions (physicochemical parameters and microbiota) encountered in cheeses.
3.2. Impact of Cheese-Making Parameters on STEC and MFGs
The different processing steps applied and the origin of the raw milk used (e.g., cow, buffalo, goat, or sheep) can influence the behavior and survival of STEC [74]. The behavior of STEC (survival, growth, or inactivation) can also be influenced by temperature and by the intrinsic physicochemical properties (pH, aw, and % lactic acid) of, and the other microflora present in, the raw milk microbiota and added starters used in different cheeses during their manufacture. At the initial stages of cheese-making, the temperature (around 30 °C) and aw value of milk provide favorable conditions for the growth of STEC and an increase in STEC level by 1–3 log CFU/g can occur [74][75]. Then, the rapid acidification (pH > 4.3) encountered during the manufacture of certain cheeses can reduce STEC cell counts by 1–4 log CFU/g, depending on the STEC serotype and the type of cheese [74][76]. Various studies have shown that when ripening is long and, therefore, the aw is low, STEC numbers decrease [74]. Nevertheless, while ripening can reduce the number of STEC cells, it cannot ensure the safety of the product if the raw milk is contaminated with STEC [77][78]. The environmental conditions during cheese processing generate stress for STEC, which can affect gene expression. Although little is known about the physiological state of STEC in cheese at different stages of production, it has been shown that the cheese-making process can trigger the production of Stx phages [79].
The various milk treatments for cheese processing also impact MFG and MFGM integrity and, consequently, the molecules involved in the MFGM–bacteria association [29][80]. Such treatments include high-temperature treatments [81][82], homogenization, and enzymatic reactions [83]. One must keep in mind that for raw milk cheeses, the curd is never heated above 54 °C. Heat treatments that kill or limit bacterial growth can also damage heat-sensitive compounds, such as glycoconjugates and associated oligosaccharides, located on the MFGM surface. Carbohydrate epitopes are well-known targets of bacterial adhesion [84]. Homogenization reduces the diameter of MFGs (ranging from 0.1 to 0.5 µm [80]) and increases the total MFGM surface area available to bacteria. Furthermore, homogenization can alter MFGM composition [82][83][85]. Treatments applied during cheese production may also alter the environment and the ability of STEC to adhere to the MFGM. In addition, physical forces applied in some cheese processing (e.g., pressed cheeses) can lead to detachment of the MFGM and their dispersion either in the product or in the whey, which will be eliminated. Nevertheless, glycoconjugates may remain intact through to end-consumption in non-pasteurized dairy products.
3.3. Location of STEC in Raw Milk Cheeses
Douëllou et al. used epifluorescence microscopy and a specific antibody coupled to FITC to assess bovine raw milk and concluded that both STEC strains assayed were localized near MFGs [86]. A natural raw milk creaming saturation assay with STEC revealed a strain-dependent tropism for the bovine raw milk cream layer and a distinct half-saturation point (8.5–9 log10 CFU.mL−1). Similar observations were presented by Brewster and Paul, suggesting that the cream layer exhibits a high capacity for E. coli O157:H7, Listeria monocytogenes, and Salmonella enterica [87]. Bacterial purification from milk by creaming seems to be not species-specific but rather general to bacteria.
The discrimination of specific bacteria in a complex product, such as raw milk cheese, is a true challenge. Furthermore, due to the modifications to milk and the MFG structure that occur during the cheese-making process, bacterial localization can vary. At the macroscopic scale, Miszczycha et al. showed that in experimentally inoculated cheeses, the levels of STEC strains belonging to serotypes O103:H2 and O145:H28 were statistically similar in the core and in the rind, regardless of the ripening conditions (traditional or industrial) [74]. The levels of serotype O26:H11 were also statistically similar in the rind. Bacterial localization in raw milk cheese has also been realized by morphologic observation on electronic micrographs [88][89][90][91] or by nonspecific DNA staining and assessment by fluorescent microscopy [88][92][93]. One study observed fluorescent mCherry-tagged Lactobacillus reuteri in raw milk [94]. However, to the authors’ knowledge, no study focusing on the localization of STEC or other pathogenic bacteria during the cheese production process has been published.
At the microscopic scale, the localization of bacteria in other dairy products, such as fermented milk, is poorly documented. Overall, few studies have focused on the localization of STEC in dairy products. More work is needed to understand STEC behavior in raw milk cheeses and their interaction with MFGs. Based on the available literature describing other bacteria [93][95][96], STEC could potentially localize near MFGs, in serum pockets, or the protein network.
References
- CNIEL Laits Liquides. Available online: https://www.filiere-laitiere.fr/fr/laits-liquides (accessed on 5 January 2022).
- International Dairy Federation. IDF Annual Report; International Dairy Federation: Schaerbeek, Belgium, 2021.
- Jost, R. Milk and Dairy Products. Ullmann’s Encycl. Ind. Chem. 2007, 23, 315–375.
- CNIEL Fromages. Available online: https://www.filiere-laitiere.fr/fr/fromages (accessed on 12 January 2022).
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The Complex Microbiota of Raw Milk. FEMS Microbiol. Rev. 2013, 37, 664–698.
- Skeie, S.B.; Håland, M.; Thorsen, I.M.; Narvhus, J.; Porcellato, D. Bulk Tank Raw Milk Microbiota Differs within and between Farms: A Moving Goalpost Challenging Quality Control. J. Dairy Sci. 2019, 102, 1959–1971.
- Machado, S.G.; Baglinière, F.; Marchand, S.; Van Coillie, E.; Vanetti, M.C.D.; De Block, J.; Heyndrickx, M. The Biodiversity of the Microbiota Producing Heat-Resistant Enzymes Responsible for Spoilage in Processed Bovine Milk and Dairy Products. Front. Microbiol. 2017, 8, 302.
- Lucey, J.A. Raw Milk Consumption: Risks and Benefits. Nutr. Today 2015, 50, 189–193.
- Crippa, G.; Zabzuni, D.; Bravi, E.; Piva, G.; De Noni, I.; Bighi, E.; Rossi, F. Randomized, Double Blind Placebo-Controlled Pilot Study of the Antihypertensive Effects of Grana Padano D.O.P. Cheese Consumption in Mild—Moderate Hypertensive Subjects. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7573–7581.
- Loss, G.; Apprich, S.; Waser, M.; Kneifel, W.; Genuneit, J.; Büchele, G.; Weber, J.; Sozanska, B.; Danielewicz, H.; Horak, E.; et al. The Protective Effect of Farm Milk Consumption on Childhood Asthma and Atopy: The GABRIELA Study. J. Allergy Clin. Immunol. 2011, 128, 766–773.e4.
- PASTURE Project Protection against Allergy: Study in Rural Environments. Available online: https://cordis.europa.eu/project/id/QLK4-CT-2001-00250/fr (accessed on 5 January 2022).
- French Ministry of Agriculture and Food Consumption of Cheeses Made from Raw Milk: Reminder of the Precautions to Take. Available online: https://agriculture.gouv.fr/consommation-de-fromages-base-de-lait-cru-rappel-des-precautions-prendre (accessed on 18 October 2021).
- Baylis, C.L. Raw Milk and Raw Milk Cheeses as Vehicles for Infection by Verotoxin-Producing Escherichia Coli. Int. J. Dairy Technol. 2009, 62, 293–307.
- Currie, A. Outbreak of Escherichia Coli O157:H7 Infections Linked to Aged Raw Milk Gouda Cheese. J. Food Prot. 2018, 81, 325–331.
- Espié, E.; Mariani-Kurkdjian, P.; Grimont, F.; Pihier, N.; Vaillant, V.; Francart, S.; Capek, I.; De Valk, H.; Vernozy-Rozand, C. Shiga-Toxin Producing Escherichia Coli O26 Infection and Unpasteurised Cows Cheese, France, 2005. In Proceedings of the 6th International Symposium on STEC, Melbourne, Australia, 30 October 2006.
- FAO; WHO. Attributing Illness Caused by Shiga Toxin-Producing Escherichia Coli (STEC) to Specific Foods; FAO: Rome, Italy, 2019.
- Honish, L.; Predy, G.; Hislop, N.; Chui, L.; Kowalewska-Grochowska, K.; Trottier, L.; Kreplin, C.; Zazulak, I. An Outbreak of E. Coli O157:H7 Hemorrhagic Colitis Associated with Unpasteurized Gouda Cheese. Can. J. Public Health 2005, 96, 182–184.
- Mungai, E.A.; Behravesh, C.; Gould, L. Increased Outbreaks Associated with Nonpasteurized Milk, United States, 2007–2012. Emerg. Infect. Dis. 2015, 21, 119–122.
- Perrin, F.; Tenenhaus-Aziza, F.; Michel, V.; Miszczycha, S.; Bel, N.; Sanaa, M. Quantitative Risk Assessment of Haemolytic and Uremic Syndrome Linked to O157:H7 and Non-O157:H7 Shiga-Toxin Producing Escherichia Coli Strains in Raw Milk Soft Cheeses. Risk Anal. 2015, 35, 109–128.
- Treacy, J. Outbreak of Shiga Toxin-Producing Escherichia Coli O157:H7 Linked to Raw Drinking Milk Resolved by Rapid Application of Advanced Pathogen Characterization Methods. Eurosurveillance 2019, 24, 1800191.
- Etcheverría, A.I.; Padola, N.L. Shiga Toxin-Producing Escherichia Coli. Virulence 2013, 4, 366–372.
- AFFSA. Bilan des Connaissances Relatives Aux Escherichia Coli Producteurs de Shiga-Toxines (STEC); French Food Safety Agency: Maisons-Alfort, France, 2003; p. 220.
- EU Directorate-General for Health and Food Safety RASFF. Available online: https://webgate.ec.europa.eu/rasff-window/screen/search (accessed on 2 February 2022).
- EFSA. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, e05926.
- EFSA. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406.
- EFSA. The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, e06971.
- Douëllou, T.; Delannoy, S.; Ganet, S.; Mariani-Kurkdjian, P.; Fach, P.; Loukiadis, E.; Montel, M.; Thevenot-Sergentet, D. Shiga Toxin-Producing Escherichia Coli Strains Isolated from Dairy Products—Genetic Diversity and Virulence Gene Profiles. Int. J. Food Microbiol. 2016, 232, 52–62.
- Claeys, W.L.; Cardoen, S.; Daube, G.; De Block, J.; Dewettinck, K.; Dierick, K.; De Zutter, L.; Huyghebaert, A.; Imberechts, H.; Thiange, P.; et al. Raw or Heated Cow Milk Consumption: Review of Risks and Benefits. Food Control 2013, 31, 251–262.
- Douëllou, T.; Montel, M.C.; Thevenot Sergentet, D. Invited Review: Anti-Adhesive Properties of Bovine Oligosaccharides and Bovine Milk Fat Globule Membrane-Associated Glycoconjugates against Bacterial Food Enteropathogens. J. Dairy Sci. 2017, 100, 3348–3359.
- Ofek, I.; Hasty, D.L.; Sharon, N. Anti-Adhesion Therapy of Bacterial Diseases: Prospects and Problems. FEMS Immunol. Med. Microbiol. 2003, 38, 181–191.
- Yoon, Y.; Lee, S.; Choi, K.-H. Microbial Benefits and Risks of Raw Milk Cheese. Food Control 2016, 63, 201–215.
- Kosmerl, E.; Rocha-Mendoza, D.; Ortega-Anaya, J.; Jiménez-Flores, R.; García-Cano, I. Improving Human Health with Milk Fat Globule Membrane, Lactic Acid Bacteria, and Bifidobacteria. Microorganisms 2021, 22, 341.
- Reinhardt, T.A.; Lippolis, J.D. Bovine Milk Fat Globule Membrane Proteome. J. Dairy Res. 2006, 73, 406–416.
- Spitsberg, V.L. Invited Review: Bovine Milk Fat Globule Membrane as a Potential Nutraceutical. J. Dairy Sci. 2005, 88, 2289–2294.
- CNIEL. L’économie Laitière en Chiffre-Edition 2021; CNIEL: Paris, France, 2021; p. 204.
- Insée Principales Caractéristiques Des Entreprises En 2017−Caractéristiques Comptables, Financières et d’emploi Des Entreprises En 2017|Insee. Available online: https://www.insee.fr/fr/statistiques/4226019?sommaire=4226092#consulter-sommaire (accessed on 10 January 2022).
- CNIEL Centre National Interprofessionnel de l’Economie Laitière. Available online: https://www.filiere-laitiere.fr/fr/ (accessed on 12 January 2022).
- Karmali, M.A.; Gannon, V.; Sargeant, J.M. Verocytotoxin-Producing Escherichia Coli (VTEC). Vet. Microbiol. 2010, 140, 360–370.
- Salaheen, S.; Kim, S.W.; Cao, H.; Wolfgang, D.R.; Hovingh, E.; Karns, J.S.; Haley, B.J.; Van Kessel, J.A.S. Antimicrobial Resistance Among Escherichia Coli Isolated from Veal Calf Operations in Pennsylvania. Foodborne Pathog. Dis. 2019, 16, 74–80.
- Brown, C.A.; Harmon, B.G.; Zhao, T.; Doyle, M.P. Experimental Escherichia Coli O157:H7 Carriage in Calves. Appl. Environ. Microbiol. 1997, 63, 27–32.
- Chapman, P.A.; Cerdán Malo, A.T.; Ellin, M.; Ashton, R.; Harkin, M.A. Escherichia Coli O157 in Cattle and Sheep at Slaughter, on Beef and Lamb Carcasses and in Raw Beef and Lamb Products in South Yorkshire, UK. Int. J. Food Microbiol. 2001, 64, 139–150.
- Sarimehmetoglu, B.; Aksoy, M.H.; Ayaz, N.D.; Ayaz, Y.; Kuplulu, O.; Kaplan, Y.Z. Detection of Escherichia Coli O157:H7 in Ground Beef Using Immunomagnetic Separation and Multiplex PCR. Food Control 2009, 20, 357–361.
- Ruegg, P.L. Practical Food Safety Interventions for Dairy Production. J. Dairy Sci. 2003, 68, E1–E9.
- WHO; FAO. Shiga Toxin-Producing Escherichia Coli (STEC) and Food: Attribution, Characterization, and Monitoring: Report; World Health Organization: Geneva, Switzerland, 2018.
- Bai, X.; Fu, S.; Zhang, J.; Fan, R.; Xu, Y.; Sun, H.; He, X.; Xu, J.; Xiong, Y. Identification and Pathogenomic Analysis of an Escherichia Coli Strain Producing a Novel Shiga Toxin 2 Subtype. Sci. Rep. 2018, 8, 6756.
- Hughes, A.C.; Zhang, Y.; Bai, X.; Xiong, Y.; Wang, Y.; Yang, X.; Xu, Q.; He, X. Structural and Functional Characterization of Stx2k, a New Subtype of Shiga Toxin 2. Microorganisms 2019, 8, 4.
- He, X.; Patfield, S.; Rasooly, R.; Mavrici, D. Novel Monoclonal Antibodies against Stx1d and 1e and Their Use for Improving Immunoassays. J. Immunol. Methods 2017, 447, 52–56.
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bover-Cid, S.; Chemaly, M.; Davies, R.; Cesare, A.D.; Herman, L.; Hilbert, F.; Lindqvist, R.; et al. Pathogenicity Assessment of Shiga Toxin-Producing Escherichia Coli (STEC) and the Public Health Risk Posed by Contamination of Food with STEC. EFSA J. 2020, 18, e05967.
- FAO/WHO STEC EXPERT GROUP Hazard Identification and Characterization: Criteria for Categorizing Shiga Toxin-Producing Escherichia Coli on a Risk Basis. J. Food Prot. 2019, 82, 7–21.
- Newton, H.J.; Sloan, J.; Bulach, D.M.; Seemann, T.; Allison, C.C.; Tauschek, M.; Robins-Browne, R.M.; Paton, J.C.; Whittam, T.S.; Paton, A.W.; et al. Shiga Toxin–Producing Escherichia Coli Strains Negative for Locus of Enterocyte Effacement. Emerg. Infect. Dis. 2009, 15, 372–380.
- Colello, R.; Krüger, A.; Velez, M.V.; Del Canto, F.; Etcheverría, A.I.; Vidal, R.; Padola, N.L. Identification and Detection of Iha Subtypes in LEE-Negative Shiga Toxin-Producing Escherichia Coli (STEC) Strains Isolated from Humans, Cattle and Food. Heliyon 2019, 5, e03015.
- Frankel, G.; Lider, O.; Hershkoviz, R.; Mould, A.P.; Kachalsky, S.G.; Candy, D.C.A.; Cahalon, L.; Humphries, M.J.; Dougan, G. The Cell-Binding Domain of Intimin from Enteropathogenic Escherichia Coli Binds to Β1 Integrins. J. Biol. Chem. 1996, 271, 20359–20364.
- Sinclair, J.F.; O’Brien, A.D. Cell Surface-Localized Nucleolin Is a Eukaryotic Receptor for the Adhesin Intimin-γ of Enterohemorrhagic Escherichia Coli O157:H7. J. Biol. Chem. 2002, 277, 2876–2885.
- Sinclair, J.F.; O’Brien, A.D. Intimin Types α, β, and γ Bind to Nucleolin with Equivalent Affinity but Lower Avidity than to the Translocated Intimin Receptor. J. Biol. Chem. 2004, 279, 33751–33758.
- Farfan, M.J.; Torres, A.G. Molecular Mechanisms That Mediate Colonization of Shiga Toxin-Producing Escherichia Coli Strains. Infect. Immun. 2012, 80, 903–913.
- Farfan, M.J.; Cantero, L.; Vidal, R.; Botkin, D.J.; Torres, A.G. Long Polar Fimbriae of Enterohemorrhagic Escherichia Coli O157:H7 Bind to Extracellular Matrix Proteins. Infect. Immun. 2011, 79, 3744–3750.
- Herold, S.; Paton, J.C.; Paton, A.W. Sab, a Novel Autotransporter of Locus of Enterocyte Effacement-Negative Shiga-Toxigenic Escherichia Coli O113:H21, Contributes to Adherence and Biofilm Formation. Infect. Immun. 2009, 77, 3234–3243.
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia Coli. Nat. Rev. Microbiol. 2004, 2, 123–140.
- McWilliams, B.D.; Torres, A.G. EHEC Adhesins. Microbiol. Spectr. 2014, 2, EHEC-0003-2013.
- Erdem, A.L.; Avelino, F.; Xicohtencatl-Cortes, J.; Girón, J.A. Host Protein Binding and Adhesive Properties of H6 and H7 Flagella of Attaching and Effacing Escherichia Coli. J. Bacteriol. 2007, 189, 7426–7435.
- Lu, Y.; Iyoda, S.; Satou, H.; Satou, H.; Itoh, K.; Saitoh, T.; Watanabe, H. A New Immunoglobulin-Binding Protein, EibG, Is Responsible for the Chain-Like Adhesion Phenotype of Locus of Enterocyte Effacement-Negative, Shiga Toxin-Producing Escherichia Coli. Infect. Immun. 2006, 74, 5747–5755.
- Rubin, D.; Zhang, W.; Karch, H.; Kuczius, T. Distinct Expression of Immunoglobulin-Binding Proteins in Shiga Toxin-Producing Escherichia Coli Implicates High Protein Stability and a Characteristic Phenotype. Toxins 2017, 9, 153.
- Jaglic, Z.; Desvaux, M.; Weiss, A.; Nesse, L.L.; Meyer, R.L.; Demnerova, K.; Schmidt, H.; Giaouris, E.; Sipailiene, A.; Teixeira, P.; et al. Surface Adhesins and Exopolymers of Selected Foodborne Pathogens. Microbiology 2014, 160, 2561–2582.
- Meynier, A.; Genot, C. Molecular and Structural Organization of Lipids in Foods: Their Fate during Digestion and Impact in Nutrition. OCL 2017, 24, D202.
- Lopez, C.; Briard-Bion, V.; Ménard, O.; Beaucher, E.; Rousseau, F.; Fauquant, J.; Leconte, N.; Robert, B. Fat Globules Selected from Whole Milk According to Their Size: Different Compositions and Structure of the Biomembrane, Revealing Sphingomyelin-Rich Domains. Food Chem. 2011, 125, 355–368.
- Raynal-Ljutovac, K.; Bouvier, J.; Gayet, C.; Simon, N.; Joffre, F.; Fine, F.; Vendeuvre, J.-L.; Lopez, C.; Chardigny, J.-M.; Michalski, M.-C.; et al. Organisation structurale et moléculaire des lipides dans les aliments: Impacts possibles sur leur digestion et leur assimilation par l’Homme. OCL 2011, 18, 324–351.
- Evers, J.M.; Haverkamp, R.G.; Holroyd, S.E.; Jameson, G.B.; Mackenzie, D.D.S.; McCarthy, O.J. Heterogeneity of Milk Fat Globule Membrane Structure and Composition as Observed Using Fluorescence Microscopy Techniques. Int. Dairy J. 2008, 18, 1081–1089.
- Keenan, T.; Mather, I. Intracellular Origin of Milk Fat Globules and the Nature of the Milk Fat Globule Membrane. In Advanced Dairy Chemistry Volume 2 Lipids; Springer: Berlin/Heidelberg, Germany, 2006; pp. 137–171.
- Lopez, C. Intracellular Origin of Milk Fat Globules, Composition and Structure of the Milk Fat Globule Membrane Highlighting the Specific Role of Sphingomyelin. In Advanced Dairy Chemistry, Volume 2: Lipids; McSweeney, P.L.H., Fox, P.F., O’Mahony, J.A., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 107–131. ISBN 978-3-030-48686-0.
- El-Zeini, H.M. Microstructure, Rheological and Geometrical Properties of Fat Globules of Milk from Different Animal Species. Pol. J. Food Nutr. Sci. 2006, 56, 147–154.
- Mather, I.H. A Review and Proposed Nomenclature for Major Proteins of the Milk-Fat Globule Membrane1,2. J. Dairy Sci. 2000, 83, 203–247.
- Farrokh, C.; Jordan, K.; Auvray, F.; Glass, K.; Oppegaard, H.; Raynaud, S.; Thevenot, D.; Condron, R.; De Reu, K.; Govaris, A.; et al. Review of Shiga-Toxin-Producing Escherichia Coli (STEC) and Their Significance in Dairy Production. Int. J. Food Microbiol. 2013, 162, 190–212.
- Rivero, M.A.; Passucci, J.A.; Rodriguez, E.M.; Parma, A.E. Role and Clinical Course of Verotoxigenic Escherichia Coli Infections in Childhood Acute Diarrhoea in Argentina. J. Med. Microbiol. 2010, 59, 345–352.
- Miszczycha, S.D.; Perrin, F.; Ganet, S.; Jamet, E.; Tenenhaus-Aziza, F.; Montel, M.-C.; Thevenot-Sergentet, D. Behavior of Different Shiga Toxin-Producing Escherichia Coli Serotypes in Various Experimentally Contaminated Raw-Milk Cheeses. Appl. Environ. Microbiol. 2013, 79, 150–158.
- Miszczycha, S.D.; Bel, N.; Gay-Perret, P.; Michel, V.; Montel, M.C.; Sergentet-Thevenot, D. Short Communication: Behavior of Different Shiga Toxin-Producing Escherichia Coli Serotypes (O26:H11, O103:H2, O145:H28, O157:H7) during the Manufacture, Ripening, and Storage of a White Mold Cheese. J. Dairy Sci. 2016, 99, 5224–5229.
- Donnelly, C. Review of Controls for Pathogen Risks in Scottish Artisan Cheeses Made from Unpasteurised Milk; Food Standards Scotland: Aberdeen, Scotland, 2018.
- Chon, J.-W.; Kim, J.-W.; Song, K.-Y.; Lim, J.-S.; Bae, D.; Kim, H.; Seo, K.-H. Fate and Survival of Listeria Monocytogenes and Escherichia Coli O157:H7 during Ripening of Cheddar Cheeses Manufactured from Unpasteurized Raw Milk. LWT 2020, 133, 109944.
- Gill, A.; Oudit, D. Enumeration of Escherichia Coli O157 in Outbreak-Associated Gouda Cheese Made with Raw Milk. J. Food Prot. 2015, 78, 1733–1737.
- Bonanno, L.; Delubac, B.; Michel, V.; Auvray, F. Influence of Stress Factors Related to Cheese-Making Process and to STEC Detection Procedure on the Induction of Stx Phages from STEC O26:H11. Front. Microbiol. 2017, 8, 296.
- Lopez, C. Focus on the Supramolecular Structure of Milk Fat in Dairy Products. Reprod. Nutr. Dev. 2005, 45, 497–511.
- Kim, H.H.; Jimenez-Flores, R. Heat-Induced Interactions between the Proteins of Milk Fat Globule Membrane and Skim Milk. J. Dairy Sci. 1995, 78, 24–35.
- Sharma, S.K.; Dalgleish, D.G. Interactions between Milk Serum Proteins and Synthetic Fat Globule Membrane during Heating of Homogenized Whole Milk. J. Agric. Food Chem. 1993, 41, 1407–1412.
- Lopez, C.; Cauty, C.; Guyomarc’h, F. Organization of Lipids in Milks, Infant Milk Formulas and Various Dairy Products: Role of Technological Processes and Potential Impacts. Dairy Sci. Technol. 2015, 95, 863–893.
- Pieters, R.J. Carbohydrate Mediated Bacterial Adhesion. In Bacterial Adhesion: Chemistry, Biology and Physics; Linke, D., Goldman, A., Eds.; Advances in Experimental Medicine and Biology; Springer: Dordrecht, The Netherlands, 2011; pp. 227–240. ISBN 978-94-007-0940-9.
- Keenan, T.W.; Dylewski, D.P.; Woodford, T.A.; Ford, R.H. Origin of Milk Fat Globules and the Nature of the Milk Fat Globule Membrane. In Developments in Dairy Chemistry—2; Springer: Dordrecht, The Netherlands, 1983; Volume 2, pp. 83–118.
- Douëllou, T.; Galia, W.; Kerangart, S.; Marchal, T.; Milhau, N.; Bastien, R.; Bouvier, M.; Buff, S.; Montel, M.-C.; Sergentet-Thevenot, D. Milk Fat Globules Hamper Adhesion of Enterohemorrhagic Escherichia Coli to Enterocytes: In Vitro and In Vivo Evidence. Front. Microbiol. 2018, 9, 947.
- Brewster, J.D.; Paul, M. Short Communication: Improved Method for Centrifugal Recovery of Bacteria from Raw Milk Applied to Sensitive Real-Time Quantitative PCR Detection of Salmonella spp. J. Dairy Sci. 2016, 99, 3375–3379.
- D’Incecco, P.; Faoro, F.; Silvetti, T.; Schrader, K.; Pellegrino, L. Mechanisms of Clostridium Tyrobutyricum Removal through Natural Creaming of Milk: A Microscopy Study. J. Dairy Sci. 2015, 98, 5164–5172.
- Laloy, E.; Vuillemard, J.-C.; El Soda, M.; Simard, R.E. Influence of the Fat Content of Cheddar Cheese on Retention and Localization of Starters. Int. Dairy J. 1996, 6, 729–740.
- Oberg, C.J.; Wr, M.; Dj, M. Microstructure of Mozzarella Cheese during Manufacture. Food Struct. 1993, 12, 251–258.
- Pitino, I.; Randazzo, C.L.; Cross, K.L.; Parker, M.L.; Bisignano, C.; Wickham, M.S.J.; Mandalari, G.; Caggia, C. Survival of Lactobacillus Rhamnosus Strains Inoculated in Cheese Matrix during Simulated Human Digestion. Food Microbiol. 2012, 31, 57–63.
- Brisson, G.; Payken, H.F.; Sharpe, J.P.; Jiménez-Flores, R. Characterization of Lactobacillus Reuteri Interaction with Milk Fat Globule Membrane Components in Dairy Products. J. Agric. Food Chem. 2010, 58, 5612–5619.
- Lopez, C.; Maillard, M.-B.; Briard-Bion, V.; Camier, B.; Hannon, J.A. Lipolysis during Ripening of Emmental Cheese Considering Organization of Fat and Preferential Localization of Bacteria. J. Agric. Food Chem. 2006, 54, 5855–5867.
- Sun, L.; Dicksved, J.; Priyashantha, H.; Lundh, Å.; Johansson, M. Distribution of Bacteria between Different Milk Fractions, Investigated Using Culture-Dependent Methods and Molecular-Based and Fluorescent Microscopy Approaches. J. Appl. Microbiol. 2019, 127, 1028–1037.
- Burdikova, Z.; Svindrych, Z.; Hickey, C.; Wilkinson, M.G.; Auty, M.A.E.; Samek, O.; Bernatova, S.; Krzyzanek, V.; Periasamy, A.; Sheehan, J.J. Application of Advanced Light Microscopic Techniques to Gain Deeper Insights into Cheese Matrix Physico-Chemistry. Dairy Sci. Technol. 2015, 95, 687–700.
- Hickey, C.D.; Sheehan, J.J.; Wilkinson, M.G.; Auty, M.A.E. Growth and Location of Bacterial Colonies within Dairy Foods Using Microscopy Techniques: A Review. Front. Microbiol. 2015, 6, 99.
More
Information
Subjects:
Microbiology; Food Science & Technology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
793
Revisions:
2 times
(View History)
Update Date:
06 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




