Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Corinne Mhiri | -- | 2884 | 2022-04-04 13:11:02 | | | |
| 2 | Dean Liu | Meta information modification | 2884 | 2022-04-06 10:59:03 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Mhiri, C.; , . Transposable Elements in Land Plants. Encyclopedia. Available online: https://encyclopedia.pub/entry/21350 (accessed on 01 March 2026).
Mhiri C, . Transposable Elements in Land Plants. Encyclopedia. Available at: https://encyclopedia.pub/entry/21350. Accessed March 01, 2026.
Mhiri, Corinne, . "Transposable Elements in Land Plants" Encyclopedia, https://encyclopedia.pub/entry/21350 (accessed March 01, 2026).
Mhiri, C., & , . (2022, April 04). Transposable Elements in Land Plants. In Encyclopedia. https://encyclopedia.pub/entry/21350
Mhiri, Corinne and . "Transposable Elements in Land Plants." Encyclopedia. Web. 04 April, 2022.
Copy Citation
Transposable elements (TEs) are important components of most plant genomes. These mobile repetitive sequences are highly diverse in terms of abundance, structure, transposition mechanisms, activity and insertion specificities across plant species.
transposable element
transposition control
plant genome
TE classification
1. Introduction
Transposable elements (TEs) can be defined as repetitive DNA sequences able to move/transpose throughout their host genome. They were first discovered in maize by Barbara McClintock in the 1940s as controlling elements able to modify gene expression and change their location upon genomic stress, such as chromosomal double-strand breaks [1]. With the development of molecular biology and sequencing technologies, the detection of mobile genetic elements has been generalized to almost all living organisms. Their high abundance in some genomes (>80% in maize) [2] and extreme diversity in transposition modes and insertion profiles resulted in a progressive interest of the research community for studying the biology of these repetitive sequences, and the way they interact and coevolve with their host genome. Initially seen as invading parasitic sequences because of their proliferative and mutational abilities [3][4], TEs have been progressively considered as important components of eukaryotic genomes since the discovery of some ‘useful’ TEs contributing to gene expression regulation or enzymatic functions [5][6]. Such opinion changes, considering TEs as ‘facilitators of evolution’ for their host organisms, have been well documented [7][8][9][10].
2. Plant TE Landscape
2.1. A Highly Variable TE Abundance
Due to their ubiquity, abundance and transposition activity, TEs have been proposed as major contributors to genome size, along with other mechanisms such as recombinational rate and polyploidization [11]. Among living organisms, land plants (regrouping Bryophytes, Pteridophytes, and seed plants), and especially flowering plants (Angiosperms), display one of the largest genome size variability exceeding 2400-fold, with C-values ranging from 0.07 pg (65 Mb/1 C) for the small carnivorous plant Genlisea tuberosa to 152.23 pg (149 Gb/1 C) for the monocot lily species Paris japonica (https://cvalues.science.kew.org/ accessed on 10 March 2022). Indeed, TEs seem to account for the variable proportion of plant genomes sequenced to date, spanning from ~3% in the small 82 Mb carnivorous Utricularia gibba [12] to ~85% in allohexaploid wheat (Triticum aestivum) [13] or maize genome [2].
2.2. Challenging Evaluation of Plant TE Diversity and Classification
Our knowledge about TEs structure, organization and transposition mechanisms has greatly progressed since the discovery of the first TE sequences in plants (the maize Ac/Ds elements) in 1984 by two different laboratories [14][15]. Transposable elements are extremely diverse and use various mechanisms to move. The so-called autonomous elements encode all specific functions to achieve their mobility, while some non-autonomous elements hitch-hike mobility proteins from autonomous copies, or from other TEs to transpose.
Several types of TEs might co-exist in a given genome, and each TE type can harbor multiple TE copies clustered into different families according to their sequence similarity. As some transposition mechanisms are prone to generate mutations, each family might have evolved over time, displaying a continuum of more or less diverged copies composed of both autonomous and defective elements [7][8][10][16]. Such behaviour made TE identification and classification a difficult task.
In 1989, the first TE nomenclature organized TEs into two classes according to their transposition intermediate [17]: (1) an RNA intermediate for retrotransposons (class I elements) that move via a replicative “copy-and-paste” mechanism, where a “mother” copy gives rise to several “daughter” copies without excising itself; (2) a DNA intermediate for DNA transposons (class II elements) that use a conservative “cut-and-paste” mechanism for their transposition, where the “mother” copy excises from its location to insert elsewhere in the genome (“jumping gene” concept).
This bimodal schematic classification has been further refined in 2007 (1) by creating subclasses in the class II group to include TEs that use a DNA replicative mechanism, such as Helitrons, and (2) by setting the “80-80-80” rule to define the identity percentage TE copies should share to belong to the same family, i.e., sequences over 80 bp sharing at least 80% sequence identity in at least 80% of their internal domain or terminal repeats (or both) [16][18][19]. This TE hierarchical classification splits TEs into the two previously cited classes (Class I and Class II), then into subclasses, orders and superfamilies [18]. This classification is based on the presence and order of coding regions for specific proteins or structural motifs present in TE sequences, and on their transposition specificities (presence and sequence of target site duplications (TSDs), i.e., short 2–11 bp DNA duplications, generated upon TE transposition at the new insertion site). For example, the presence of long terminal repeats (LTRs) in direct orientation at TE 3′ and 5′ ends is the signature of LTR-retrotransposons (LTR-RTs). The occurrence of the reverse transcriptase (RT) domain in TE ORF is a hallmark of most but not all Class I retrotransposons. Like self-replicating entities as viruses, TEs have a modular evolution, exchanging essential or facultative protein-coding domains that may blur TE classification [19].
If these first four classification levels are generally well accepted, the need of extra levels to reach the TE family level is still unclear and may vary according to the TE considered. For example, the superfamily subgroups “chromovirus” or “non-chromovirus” have been introduced for Gypsy LTR-RTs [20]. An increase in family/lineage number or even new superfamilies will probably arise with the accumulation of genome sequencing data and the improvement of bioinformatic pipelines for TE detection and annotation. This hierarchical classification also does not include some non-autonomous TEs, i.e., TEs that are still able to transpose but need extra-function in trans coming from another TE element (phylogenetically related or not). This includes LARDs (large retrotransposon derivatives), TRIMs (transposon in miniature) and SMARTs (small LTR-retrotransposon) for Class I TEs, and MITEs (miniature inverted-repeats transposable elements), SNACs (small non-autonomous CACTA) or MULEs (Mu-like elements) for class II TEs. Many such non-autonomous elements share enough sequence similarity/motifs to be easily linked to a candidate “helper” autonomous TE family, as reported in rice with the isolation of the complete RIRE2 Gyspy retrotransposon displaying a high LTR sequence similarity to sequence extremities of the defective Dasheng retrotransposon [21]. Another example is the description of both complete and truncated—but nevertheless active—CACTA Caspar elements in Triticeae [16][22]. Some other non-autonomous TEs may not present any clear feature allowing them to be classified, as some non-autonomous short TIR-harboring Tes that may share only a few bases homology with the autonomous helper [16]. In the classification we propose, we choose to include the non-autonomous SINES as a full order, as these TEs do not correspond to deleted versions of autonomous class I TEs.
Table 1 and Figure 1 present, respectively, the up-to-date classification and structures of plant transposable elements adapted from Wicker et al. [16]. It is important to note that only a subset of existing TE superfamilies in all living organisms (as reported in repbase https://www.girinst.org/repbase/update/browse.php accessed on 10 March 2022) has been detected in land plants (~30% of class II superfamilies, and ~17% of class I—representing 20% of all described superfamilies). Plant TEs fall into six different orders, four orders corresponding to class I elements (LTR-RTs, Penelope-like elements (PLE), long interspaced nuclear elements (LINEs), short interspaced nuclear elements (SINEs)) and two orders including elements of class II (terminal inverted repeat (TIR) transposons, Helitrons) (Table 1).
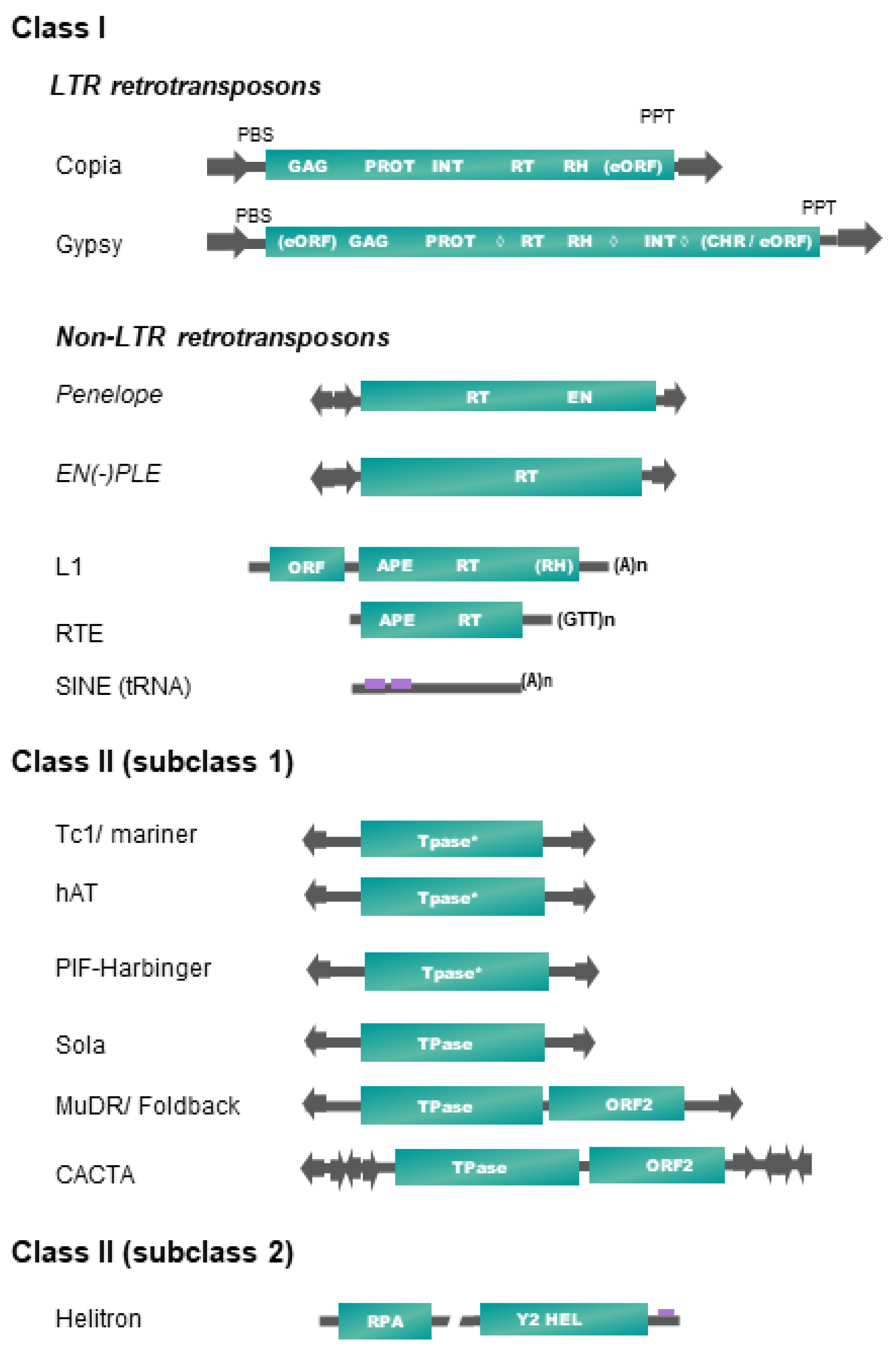
Figure 1. Structure and organization of plant transposable element superfamilies (adapted from [16]). Schemes are not to scale. Protein coding domains: APE = apurinic endonuclease, CHR = chromodomain, EN = endonuclease, GAG = capsid protein, HEL = helicase, INT = integrase, PROT = proteinase, RH = RNAse H, RPA = replication protein A, RT = reverse transcriptase. eORF = extra open reading frame (unknown function), Tpase = transposase (* with DDE motif), YR = tyrosine recombinase, Y2 = YR with YY motif, ◊ = different possible locations of an additional cellular-like ribonuclease H (aRH) specific of the Tat lineages (see Table 1). Optional protein-coding domains only present in some superfamily lineages are indicated in brackets. Some structural features are also represented. Terminal repeats in the same or reverse orientation are indicated by black arrows, and purple rectangles refer to diagnostic sequences present in non-coding sequences. Specific base termination of some TEs are also indicated. PBS = primer binding site, PPT = poly purine tract. Interrupted line in Helitron representation means that the region may contain one or more additional ORFs.
Table 1. Plant transposable element (TE) classification compiled from [16] with updates from [23] for Copia lineages, ref. [20] for Gypsy LTR retrotransposons (LTR-RTs), ref. [24][25][26] for Penelope-like elements (PLEs), ref. [27] for long interspaced nuclear elements (LINEs), ref. [28][29] for short interspaced nuclear elements (SINEs), and [30][31] for Sola elements.
| Class | Order (Non-Autonomous TE Name) |
Superfamily | Family/Lineage | Plant Family Examples |
|---|---|---|---|---|
| Class I | LTR-Retrotransposons | Copia | Osser | Volvox canteri Osser |
| (retrotransposons) | (LARD) | Bryco | representatives in moss species | |
| (TRIM/SMART) | Lyco | representatives in clubmosses species (Lycopodiaceae) | ||
| Gymco-I | representatives in gymnosperms species | |||
| Gymco-II | representatives in gymnosperms species | |||
| Gymco-III | representatives in gymnosperms species | |||
| Gymco-IV | representatives in gymnosperms species | |||
| Ale/Retrofit | Oryza longistaminata Retrofit, Oryza sativa Hopscotch | |||
| Ivana | Oryza sativa Oryko1-1 and Ilona, Hordeum vulgare HORPIA, Nicotiana tabacum Queenti | |||
| Ikeros | Zea mays Sto-4 | |||
| Tork | Nicotiana tabacum Tnt1, Tto1 and Tnt2, Solanum lycopersicum Tork4, Ipomea batatas Batata | |||
| Alesia | low copy number representatives in many Angiosperms, close to the Ale lineage | |||
| Angela | Triticum aestivum Angela, Oryza sativa RIRE1, Hordeum vulgare BARE1 | |||
| Bianca | Triticeae Bianca, Arabidopsis thaliana RomaniAT5 | |||
| SIRE/Maximus | Solanum lycopersicum ToRTL1, Zea mays Opie-2, Glycine max SIRE1 | |||
| TAR | Oryza spp. Houba and Osr-1, Arabidopsis thaliana ATcopia95 | |||
| Gypsy (Chromovirus) | Galadriel | Solanum esculentum Galadriel, Musa Monkey, Tntom1 | ||
| Tekay | Hordeum vulgare Bagy-1, Arabidopsis thaliana Legolas Peabody, Oryza sativa RIRE3, Lilium henryi Del | |||
| Reina | Zea mays Reina, Arabidopsis thaliana Gloin or Gimli | |||
| CRM | Zea mays CRM (centromeric retrotransposon of maize), Beta vulgaris Beetle1, Oryza sativa RIRE7 | |||
| (Non-chromovirus) | Phygy | Phycomitrella patens Chr21 (4035670,4045566) | ||
| Selgy | Selaginella moellendorffii LTR-RT | |||
| Athila | Arabidopsis thaliana Athila4-1, Diaspora, Hordeum vulgare Bagy-2 | |||
| TatI | Selaginella moellendorffii LTR-RT | |||
| TatII | Picea abies, Picea glauca LTR-RTs | |||
| TatIII | Picea abies, Picea glauca LTR-RTs | |||
| Ogre/TatIV + TatV | Pisum sativum Ogre | |||
| Retand/TatVI | Zea mays Cinful-1, Arabidopsis thaliana Tat4-1, Oryza sativa RIRE2, Sorghum bicolor RetroSor1, Silene latifolia Retand |
|||
| Non-LTR retrotransposons PLE |
Penelope/Poseidon | Pinus taeda (loblolly pine) and Picea abies (Norway spruce) Dryad PLEs by horizontal transfer | ||
| EN(-)PLE | Selaginella moellendorffii spike moss, Pinus taeda and Picea abies EN(-)PLEs | |||
| LINE | L1 | Llb | sweet potato Llb, Beta vulgaris BvL1 | |
| LINE-CS | Cannabis sativa LINE-CS, Beta vulgaris Belline2, Belline5 | |||
| BNR | Beta vulgaris Belline1/BNR | |||
| PUR | Carica papaya L1-26_Cpa, Solanum tuberosum L1-3_Stu, Vitis vinifera | |||
| Cin4 | Zea mays Cin4 | |||
| Karma | Oryza sativa Karma | |||
| nubo | Oryza sativa LINE-1 or OSLINE1-4, Zea mays L1-2_ZM | |||
| RTE | plant RTE | Malus x domestica RTE-1_Mad, Solanum tuberosum RTE-1_Stu | ||
| SINE | tRNA | Nicotiana tabacum TS, Au, Solanales SolS-II, Brassicale BraS-I, SB families, mainly found in Angiosperm | ||
| Class II Subclass 1 |
TIR (MITE) | Tc1-Mariner | Stowaway (MITE): Sorghum bicolor Stowaway, Brassica BraSto | |
| hAT | Zea mays Ac/Ds, Antirrhinum majus Tam3, Nicotiana tabacum Slide | |||
| Sola | Physcomitrella Sola1, found also in Capsicum annuum and C. baccatum | |||
| (MULE) | MuDR-Foldback | Zea mays Mu, MULEs | ||
| PIF-Harbinger | Zea mays PIFa, Oryza sativa Pong; Tourist (MITE): mPing/Ping; mPIF/PIFa | |||
| CACTA | Zea mays En/Spm, Arabidopsis thaliana CAC1, Antirrhinum majus Tam1, Petunia hybrida PsI | |||
| Subclass 2 | Helitron | Helitron | Oryza sativa, Arabidopsis thaliana AthE1 Atrep, Ipomoea tricolor Hel-It1 |
In plants, class II elements (subclass 1) belonging to the TIR order (also called DNA transposons) fall into six superfamilies, based on the structure and sequence of their transposase and on the sequence of their terminal inverted repeats (TIRs) (Figure 1). Transposase is the protein catalyzing their transposition, while TIRs harbor key sequences recognized by the DNA-binding domains of the transposase during a transposition event. Some TIR elements also harbor additional coding sequences, as the maize MuDR, and plant CACTA or PIF/Harbinger elements [16]. Most of these superfamilies are also characterized by specific target site duplications (TSDs) lengths, generated after the filling of DNA nicks generated by the transposase on the integration site. Their transposition is not always strictly conservative and could lead to an increase of copy number if it occurs before a DNA replication fork [32].
Replicating plant TEs fall into two major groups: (1) Helitrons (class II-subclass 2, see Table 1) replicate through a rolling-circle (RC) mechanism from one DNA strand, without generating TSDs, by using a RepHel protein with a RC replication initiator (Rep) and DNA helicase (Hel) domains, in association with an ssDNA-binding “replication protein A” (RPA) [33]. (2) Class I retrotransposons replicate from RNA templates by reverse transcription using a TE-encoded reverse transcriptase (RT) and use at least one additional protein to mediate their insertion into their host genome, such as endonuclease (EN) or DDE integrase (INT). We do not include the DIRS superfamily as a member of land plants, as DIRS elements have only been found in green algae until now [34].
Among the four retrotransposon orders present in plants, SINEs occupy a particular place, as these small non-coding and non-autonomous elements of a few hundred base pairs exploit the transposition machinery of LINEs to ensure their amplification. Plant SINEs are derived from tRNAs [28]. They are transcribed by polymerase III, harbor short degenerated internal promoters (A and B boxes), and display mostly A tail at the 3′-end. Apart from these small structural domains, SINEs display a high sequence diversity that hinders their detection and characterization. Recently, a 37 pb Angio-domain located in the 3′-end has been reported in many Angiosperm SINEs [29].
The second non-LTR retrotransposon order present in plants, LINEs, contains elements belonging to the L1 and the RTE (retrotransposable element) superfamilies (Table 1 and Figure 1), which are two of the five known superfamilies of LINEs detected in eukaryotes [16]. RTE and L1 LINEs have one or two open reading frames (ORFs), respectively, and code for proteins required for retrotransposition, such as an endonuclease (EN), a RT, and often a ribonuclease H (RNase H (RH)). The L1 ORF1 is involved in the binding, protection and transport of the RNA intermediate used for retrotransposition. At their 3′-end lies a stretch of (A)n for L1 or (GTT)n for RTE involved in the reverse transcription initiation. A recent study shows that plant LINEs extracted from 23 genomes fall into only seven L1 and one RTE families/lineages/subclades [27]. As the reverse transcription starts from the 3′-end of LINEs and does not always reach the 5′-end, many incomplete daughter copies can be generated.
Between their bordering direct repeats 5′-LTR and 3′-LTR, autonomous LTR-retrotransposons (LTR-RTs) code for structural capsid-like (GAG) and functional (POL) proteins needed for their retrotransposition cycle (RT = reverse transcriptase, RH, INT = integrase), resembling the replication cycle of retroviruses. Only two out of the five superfamilies found in eukaryotes are represented in plants [16]. Plant LTR-RTs are further classified into Copia/Ty1 or Gypsy/Ty3 superfamilies according to the order of their coding pol domains. Recently, a systematic survey of plant LTR-RTs in 80 plant genomes refined the classification by introducing 16 lineages/families into the Copia/Ty1 superfamily and 14 lineages/families into the Gypsy/Ty3 group (six with a chromo-domain and eight without) [20]. Two Gypsy lineages, Chlamyvir and Tcn1, having only representatives in algae and non-Viridiplantae species, have not been included in Table 1. Non-autonomous derivatives of variable size (from a few hundred bp up to 25 kb) have been characterized in plants, containing between both LTRs a DNA sequence of variable length, either non-coding or reminiscent of some retrotransposon internal domains. Large internal sequences (>4 kb) define LARDs (large retrotransposon derivatives), and short ones (<4 kb) are often called TRIM (terminal repeat retrotransposon in miniature) [16].
Retrotransposons belonging to the Penelope-like elements (PLE) order are also found in some plant genomes, but with a patchy distribution. PLE encode an RT domain related to telomerase, a highly specialized class of non-mobile RTs responsible for chromosome end maintenance in most eukaryotes. Some PLEs also carry a second EN domain with a specific GIY-YIG motif. PLEs are bordered by repeats in direct or reverse orientation and are often subjected to 5′ truncation upon retrotransposition, as non-LTR retrotransposons. EN(+)PLEs (Dryads elements belonging to the Penelope/Poseidon group) and EN(-)PLEs have been found in some Conifer genomes (Table 1, Figure 1), and some of them were presumably derived from a horizontal transfer (HT) event [26].
The accumulation of sequenced genomes and TE detection pipelines allow the analysis and comparison of TE composition and diversity across plant genera. Figure 2 presents a heatmap of genomic percentage of four types of TEs—LTR-RTs, LINEs, SINEs and TIR DNA transposons—across 74 Angiosperm species displaying variable genome sizes (Data collected from [35] in the Supplementary Tables S1 and S2 of this article). Among the different types of TEs, LTR-RTs (Copia/Ty1 and Gypsy/Ty3) occupy the largest proportion of these genomes, the highest being up to 80% in Zea mays. Such an increase can result from rapid amplification of only a few families. For example, Oryza australiensis has undergone a recent burst of transposition involving only three families (one Copia = RIRE1 and two Gypsy = Wallabi and Kangourou), which compose 60% of its genome [36]. Genomes of the legume tribe Fabeae are also dominated by the Ty3/Gypsy Ogre family/lineage, that accounts for 57% of genome size variation on average in this clade [37]. The predominance of one type of LTR-RT may vary depending on the taxa considered. For example, in Gossypium species, Copia LTR-RTs have accumulated in the small genome of G. raimondii (880 Mb), while Gypsy LTR-RTs (mainly Gorge3) have proliferated in large genomes lineages of G. herbaceum 1667 Mb) and G. exiguum (2460 Mb) [38]. Plant species belonging to the same order (see Brassicales, Poales, Figure 2) might display different TE compositions and genome sizes. Some plant species also harbor specific TE composition as shown in Figure 2, with dominance of LINEs retrotransposons or TIR DNA transposon for the aquatic plant coontail Ceratophyllum demersum (33.6% of total genome size) and the small herbaceous plant Trichopus zeylanicus (~27.3% of total genome size). Non-LTR-retrotransposons have been shown to be abundant (~11.7%) in other plant genomes, such as Arachis ipaenis, one of the peanut parental genomes [39].
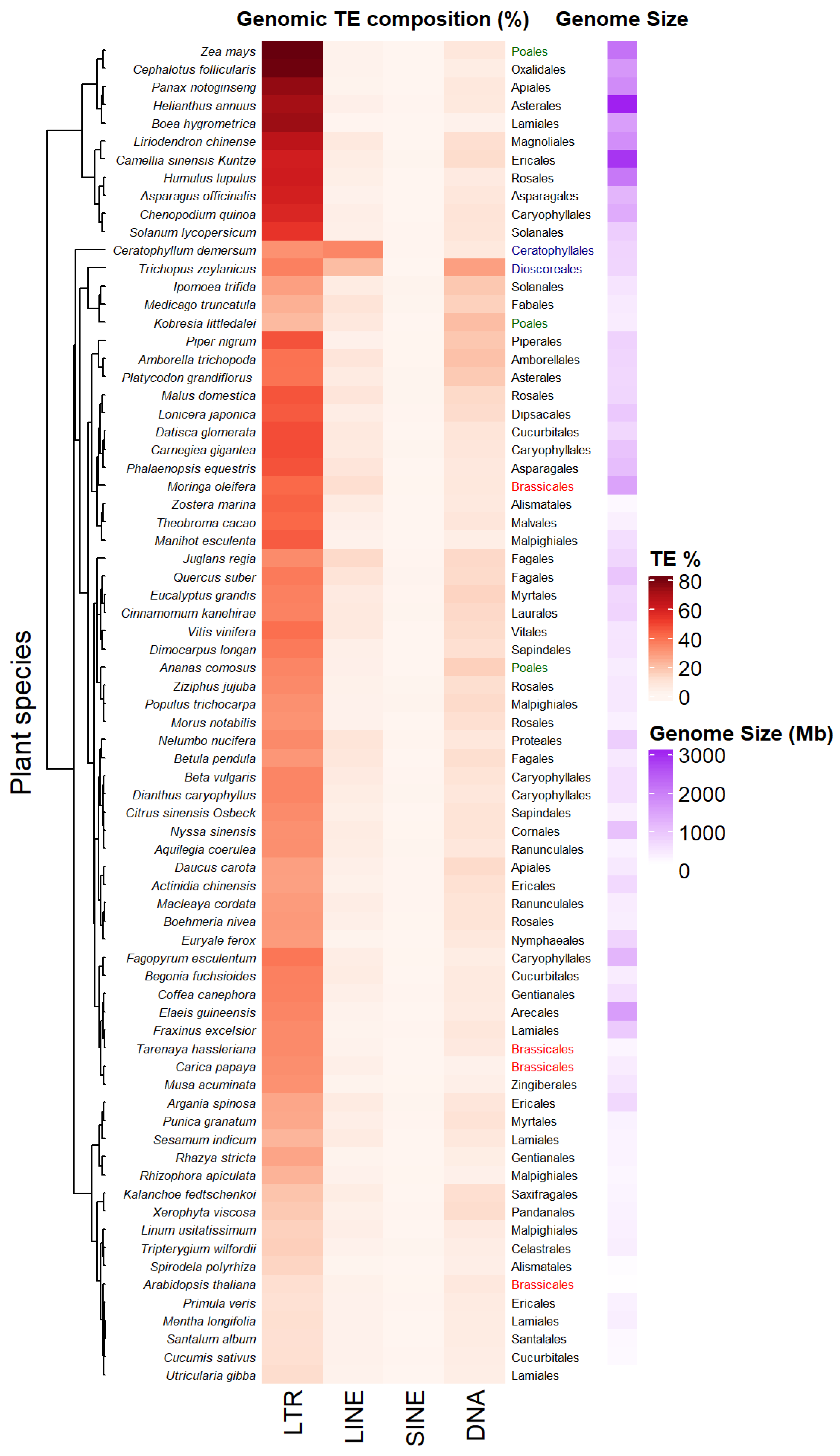
Figure 2. Transposable elements (TE) profiles in some land plant genomes. Species are clustered according to their TE profiles. TE percentages, plant orders and genome size estimations from 74 land plant species have been collected from [35] (data collected in Supporting information, Tables S1 and S2 from [35]). Some plant orders as Poales or Brassicales have been highlighted in colors (green and red respectively) in order to underline the diversity of TE composition between species belonging to the same plant order. Plant belonging to different orders as Dioscoreales and Ceratophyllales (in blue) can share close TE composition.
References
- McClintock, B. Mutable Loci in Maize. Annu. Rep. Dir. Dep. Genet. Carnegie Inst. Wash. 1951, 50, 174–181.
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 Maize Genome: Complexity, Diversity, and Dynamics. Science 2009, 326, 1112–1115.
- Doolittle, W.F.; Sapienza, C. Selfish Genes, the Phenotype Paradigm and Genome Evolution. Nature 1980, 284, 601–603.
- Orgel, L.E.; Crick, F.H.C. Selfish DNA: The Ultimate Parasite. Nature 1980, 284, 604–607.
- Oliver, K.R.; Greene, W.K. Transposable Elements: Powerful Facilitators of Evolution. BioEssays 2009, 31, 703–714.
- Kazazian, H.H. Mobile Elements: Drivers of Genome Evolution. Science 2004, 303, 1626–1632.
- Biémont, C.; Vieira, C. Junk DNA as an Evolutionary Force. Nature 2006, 443, 521–524.
- Hua-Van, A.; Le Rouzic, A.; Boutin, T.S.; Filée, J.; Capy, P. The Struggle for Life of the Genome’s Selfish Architects. Biol. Direct 2011, 6, 19.
- Lisch, D. How Important Are Transposons for Plant Evolution? Nat. Rev. Genet. 2013, 14, 49–61.
- Schulman, A.H.; Wicker, T. A Field Guide to Transposable Elements. In Plant Transposons and Genome Dynamics in Evolution; Fedoroff, N.V., Ed.; Wiley-Blackwell: Oxford, UK, 2013; pp. 15–40. ISBN 978-1-118-50015-6.
- Bennetzen, J.L.; Wang, H. The Contributions of Transposable Elements to the Structure, Function, and Evolution of Plant Genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530.
- Ibarra-Laclette, E.; Lyons, E.; Hernández-Guzmán, G.; Pérez-Torres, C.A.; Carretero-Paulet, L.; Chang, T.-H.; Lan, T.; Welch, A.J.; Juárez, M.J.A.; Simpson, J.; et al. Architecture and Evolution of a Minute Plant Genome. Nature 2013, 498, 94–98.
- Wicker, T.; Gundlach, H.; Spannagl, M.; Uauy, C.; Borrill, P.; Ramírez-González, R.H.; De Oliveira, R.; International Wheat Genome Sequencing Consortium; Mayer, K.F.X.; Paux, E.; et al. Impact of Transposable Elements on Genome Structure and Evolution in Bread Wheat. Genome Biol. 2018, 19, 103.
- Döring, H.P.; Tillmann, E.; Starlinger, P. DNA Sequence of the Maize Transposable Element Dissociation. Nature 1984, 307, 127–130.
- Pohlman, R.F.; Fedoroff, N.V.; Messing, J. The Nucleotide Sequence of the Maize Controlling Element Activator. Cell 1984, 37, 635–643.
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A Unified Classification System for Eukaryotic Transposable Elements. Nat. Rev. Genet. 2007, 8, 973–982.
- Finnegan, D.J. Eukaryotic Transposable Elements and Genome Evolution. Trends Genet. 1989, 5, 103–107.
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten Things You Should Know about Transposable Elements. Genome Biol. 2018, 19, 199.
- Wells, J.N.; Feschotte, C. A Field Guide to Eukaryotic Transposable Elements. Annu. Rev. Genet. 2020, 54, 539–561.
- Neumann, P.; Novák, P.; Hoštáková, N.; Macas, J. Systematic Survey of Plant LTR-Retrotransposons Elucidates Phylogenetic Relationships of Their Polyprotein Domains and Provides a Reference for Element Classification. Mob. DNA 2019, 10, 1.
- Jiang, N.; Jordan, I.K.; Wessler, S.R. Dasheng and RIRE2. A Nonautonomous Long Terminal Repeat Element and Its Putative Autonomous Partner in the Rice Genome. Plant Physiol. 2002, 130, 1697–1705.
- Wicker, T.; Guyot, R.; Yahiaoui, N.; Keller, B. CACTA Transposons in Triticeae. A Diverse Family of High-Copy Repetitive Elements. Plant Physiol. 2003, 132, 52–63.
- Stritt, C.; Thieme, M.; Roulin, A.C. Rare Transposable Elements Challenge the Prevailing View of Transposition Dynamics in Plants. Am. J. Bot. 2021, 108, 1310–1314.
- Arkhipova, I.R. Distribution and Phylogeny of Penelope-Like Elements in Eukaryotes. Syst. Biol. 2006, 55, 875–885.
- Craig, R.J.; Yushenova, I.A.; Rodriguez, F.; Arkhipova, I.R. An Ancient Clade of Penelope-Like Retroelements with Permuted Domains Is Present in the Green Lineage and Protists, and Dominates Many Invertebrate Genomes. Mol. Biol. Evol. 2021, 38, 5005–5020.
- Lin, X.; Faridi, N.; Casola, C. An Ancient Transkingdom Horizontal Transfer of Penelope -like Retroelements from Arthropods to Conifers. Genome Biol. Evol. 2016, 8, 1252–1266.
- Heitkam, T.; Holtgräwe, D.; Dohm, J.C.; Minoche, A.E.; Himmelbauer, H.; Weisshaar, B.; Schmidt, T. Profiling of Extensively Diversified Plant LINEs Reveals Distinct Plant-Specific Subclades. Plant J. 2014, 79, 385–397.
- Wenke, T.; Döbel, T.; Sörensen, T.R.; Junghans, H.; Weisshaar, B.; Schmidt, T. Targeted Identification of Short Interspersed Nuclear Element Families Shows Their Widespread Existence and Extreme Heterogeneity in Plant Genomes. Plant Cell 2011, 23, 3117–3128.
- Seibt, K.M.; Schmidt, T.; Heitkam, T. The Conserved 3′ Angio-domain Defines a Superfamily of Short Interspersed Nuclear Elements (SINEs) in Higher Plants. Plant J. 2020, 101, 681–699.
- Bao, W.; Jurka, M.G.; Kapitonov, V.V.; Jurka, J. New Superfamilies of Eukaryotic DNA Transposons and Their Internal Divisions. Mol. Biol. Evol. 2009, 26, 983–993.
- De Assis, R.; Baba, V.Y.; Cintra, L.A.; Gonçalves, L.S.A.; Rodrigues, R.; Vanzela, A.L.L. Genome Relationships and LTR-Retrotransposon Diversity in Three Cultivated Capsicum L. (Solanaceae) Species. BMC Genom. 2020, 21, 237.
- Feschotte, C.; Pritham, E.J. DNA Transposons and the Evolution of Eukaryotic Genomes. Annu. Rev. Genet. 2007, 41, 331–368.
- Kapitonov, V.V.; Jurka, J. Helitrons on a Roll: Eukaryotic Rolling-Circle Transposons. Trends Genet. 2007, 23, 521–529.
- Piednoël, M.; Gonçalves, I.R.; Higuet, D.; Bonnivard, E. Eukaryote DIRS1-like Retrotransposons: An Overview. BMC Genom. 2011, 12, 621.
- Wang, D.; Zheng, Z.; Li, Y.; Hu, H.; Wang, Z.; Du, X.; Zhang, S.; Zhu, M.; Dong, L.; Ren, G.; et al. Which Factors Contribute Most to Genome Size Variation within Angiosperms? Ecol. Evol. 2021, 11, 2660–2668.
- Piegu, B.; Guyot, R.; Picault, N.; Roulin, A.; Saniyal, A.; Kim, H.; Collura, K.; Brar, D.S.; Jackson, S.; Wing, R.A.; et al. Doubling Genome Size without Polyploidization: Dynamics of Retrotransposition-Driven Genomic Expansions in Oryza Australiensis, a Wild Relative of Rice. Genome Res. 2006, 16, 1262–1269.
- Macas, J.; Novák, P.; Pellicer, J.; Čížková, J.; Koblížková, A.; Neumann, P.; Fuková, I.; Doležel, J.; Kelly, L.J.; Leitch, I.J. In Depth Characterization of Repetitive DNA in 23 Plant Genomes Reveals Sources of Genome Size Variation in the Legume Tribe Fabeae. PLoS ONE 2015, 10, e0143424.
- Hawkins, J.S.; Kim, H.; Nason, J.D.; Wing, R.A.; Wendel, J.F. Differential Lineage-Specific Amplification of Transposable Elements Is Responsible for Genome Size Variation in Gossypium. Genome Res. 2006, 16, 1252–1261.
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The Genome Sequences of Arachis Duranensis and Arachis Ipaensis, the Diploid Ancestors of Cultivated Peanut. Nat. Genet. 2016, 48, 438–446.
More
Information
Subjects:
Evolutionary Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.0K
Revisions:
2 times
(View History)
Update Date:
06 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





