Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jing Ma | -- | 2258 | 2022-04-02 08:18:16 | | | |
| 2 | Catherine Yang | Meta information modification | 2258 | 2022-04-06 04:28:10 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Ma, J.; Cheng, Y.; Zhong, P.; , .; Aung, M.; Lizhuang, H.; Zhu, W. Hydrogenosome. Encyclopedia. Available online: https://encyclopedia.pub/entry/21306 (accessed on 07 February 2026).
Ma J, Cheng Y, Zhong P, , Aung M, Lizhuang H, et al. Hydrogenosome. Encyclopedia. Available at: https://encyclopedia.pub/entry/21306. Accessed February 07, 2026.
Ma, Jing, Yanfen Cheng, Pei Zhong, , Min Aung, Hao Lizhuang, Weiyun Zhu. "Hydrogenosome" Encyclopedia, https://encyclopedia.pub/entry/21306 (accessed February 07, 2026).
Ma, J., Cheng, Y., Zhong, P., , ., Aung, M., Lizhuang, H., & Zhu, W. (2022, April 02). Hydrogenosome. In Encyclopedia. https://encyclopedia.pub/entry/21306
Ma, Jing, et al. "Hydrogenosome." Encyclopedia. Web. 02 April, 2022.
Copy Citation
Hydrogenosome is a kind of membrane-bound organelle that widely exists in some evolutionarily distant protozoa and fungi, such as trichomonas, anaerobic fungi, endoamoeba and microsporidia. These microorganisms are anaerobic or microanaerobic, and they do not have mitochondria. Instead, they rely on the hydrogenosome, a kind of mitochondrion-related organelles (MROs), to metabolize organic matter under anoxic conditions, producing ATP to maintain their metabolism and growth.
hydrogenosome
anaerobic fungi
1. The Origin of the Hydrogenosome
Two billion years ago, the concentration of oxygen (O2) in the atmosphere rose sharply because of the photosynthesis of ocean surface cyanobacteria, which changed the proportion of the main components in the atmosphere. The transition from an anaerobic to an aerobic environment had a serious impact on the survival of organisms. In an aerobic environment, some aerobic bacteria (such as α-proteobacteria) can utilize organic matter and use O2 as an electron acceptor to produce ATP for energy through the process of tricarboxylic acid cycle (TCA) and electron transport. After being swallowed by an amitochondriate eukaryotic host cell or an archaeal cell [1], these ancient aerobic bacteria may have evolved into respiratory organelles, known as mitochondrion, in a long-term symbiotic relationship. Mitochondria are important sites for O2 utilization, energy production, material conversion, and metabolic regulation, and it has been thought as the organelles common to eukaryotic cells for a long time. However, since the 1980s, researchers have found that there are some obligatory or facultative anaerobic protozoa and fungi, such as trichomonas, anaerobic fungi, amoeba, and microsporidia that do not contain mitochondria, but a kind of MROs, including hydrogenosome, mitosome, and so forth, which can transfer electrons in other ways and release ATP to supply energy for cells.
The question about the origin of the hydrogenosome has puzzled many scientists. The presence of hydrogenosome in diverse evolutionarily distant organisms (Figure 1) indicates that it has evolved independently on several occasions.
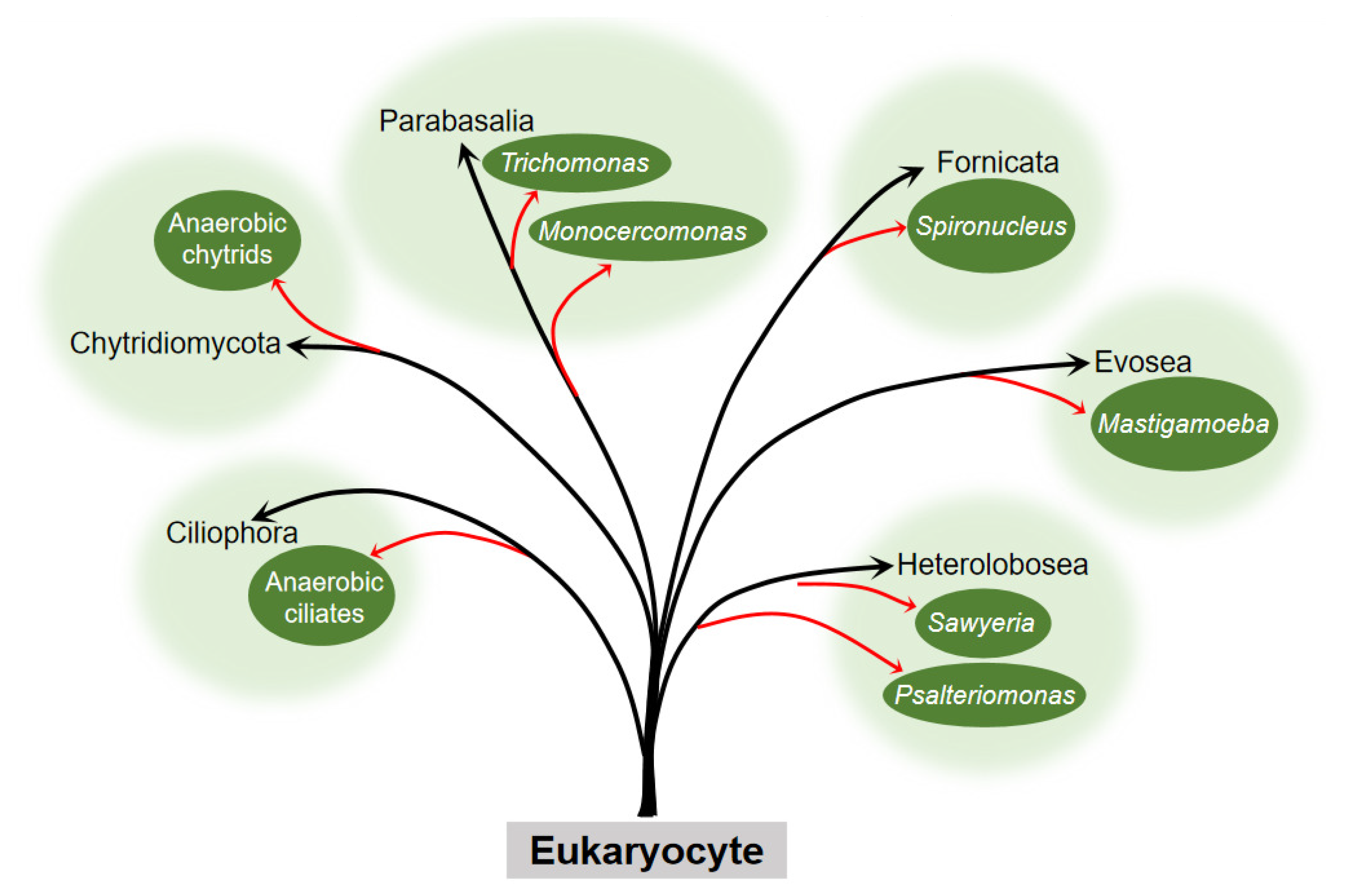
Figure 1. Occurrence of the hydrogenosome within the six eukaryocyte supergroups. Embranchment reflects the taxons of eukaryocyte with the hydrogenosome. The black arrow points to the phylum, and the red arrow points to the clades or genus to which the eukaryocyte belongs. Branch length is not to scale.
Earlier, no organellar DNA was detected or isolated from the hydrogenosome, which hampered the direct genetic demonstration of its evolutionary ancestry. Until 2005, phylogenetic analysis of the integrant hydrogenosomal gene of Nyctotherus ovalis provided explicit evidence that the hydrogenosome is indeed a modified mitochondrion [2]. However, since the enzymes and metabolism progress of the hydrogenosome in different organisms are not exactly the same, and there is no complete series of evidence to elucidate its evolution, different hypotheses about the origin of the hydrogenosome exist. These hypotheses can ultimately be generalized or covered in two arguments, whether the hydrogenosome is a degraded form of mitochondria that lost some functional proteins and gene fragments to adapt to an anaerobic environment, or the two organelles originate from the same or different endosymbionts to cope with different atmospheric conditions and energy driving forces (Figure 2).
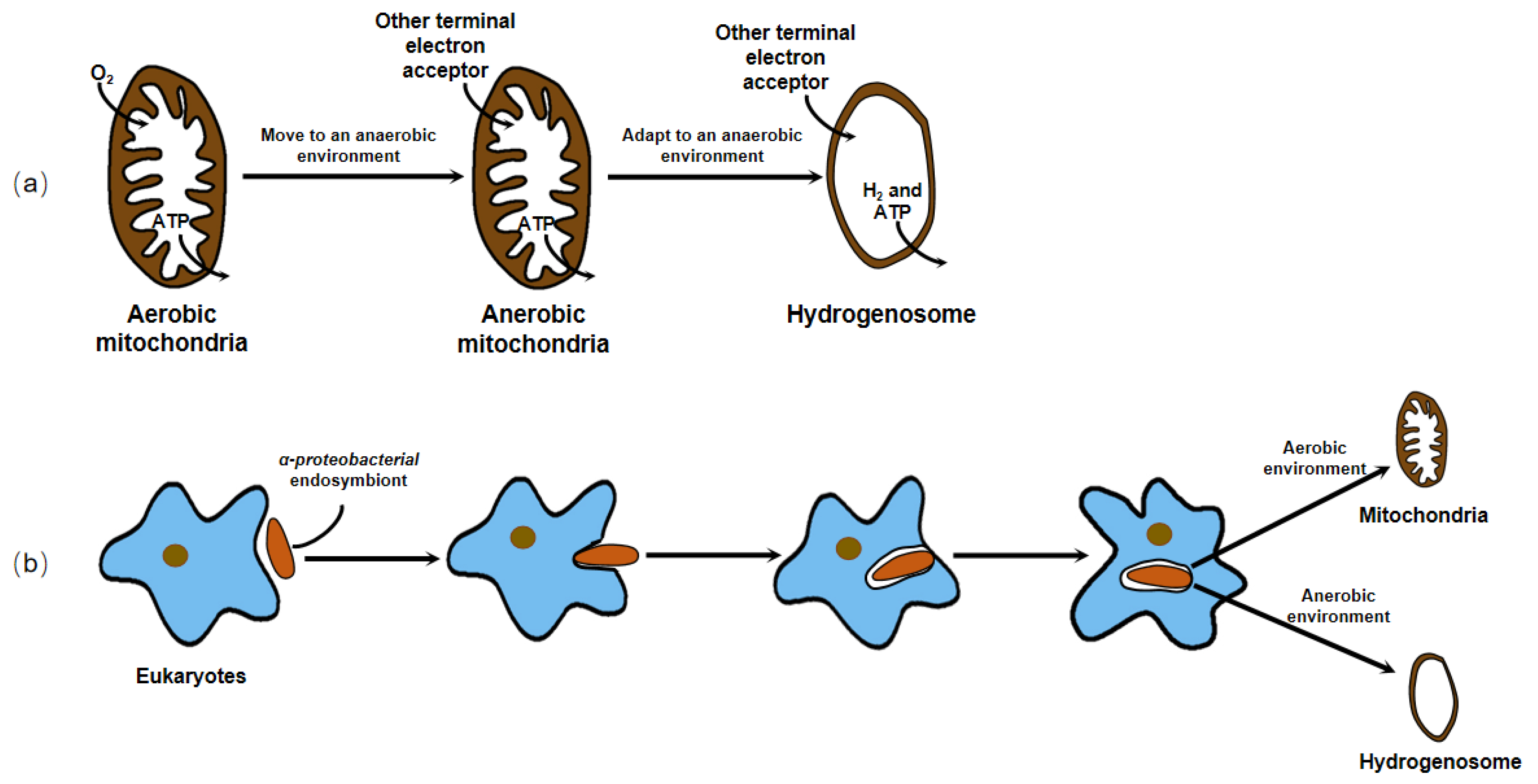
Figure 2. There are two dominant hypotheses about the origin of the hydrogenosome. (a) The hydrogenosome is a degraded form of mitochondria that lost some functional proteins and gene fragments to adapt to an anaerobic environment. (b) The hydrogenosome and mitochondria originate from the same or different endosymbionts to cope with different atmospheric conditions and energy driving forces.
For decades, researchers have conducted a large number of experiments using molecular biology, molecular genetics, and systematic taxonomy to find out the origin of the hydrogenosome. There is a large body of evidence, for example, their clearly semblable appearance [3], their closely related enzymes with important functions [4][5][6], and analogous organelle gene sequences [2][7], which support both hypotheses. Nowadays, most researchers who work on the hydrogenosomes agree with the view that these organelles originated from mitochondrion, losing partial or all mitochondrial genome in anaerobic environments.
2. Structure and Function of the Hydrogenosome
2.1. Structure of the Hydrogenosome
Most hydrogenosomes are spherical or slightly elongated granules, with a homogeneous particle matrix inside. However, in some cells, the hydrogenosomes are not spherical but are very elongated, such as those found in Monocercomonas sp. [8]. The hydrogenosomes found so far are encased by two adjacent membranes, which are very thin, presenting a thickness of only a few nanometers [9]. Although researchers have reported that the hydrogenosome of Neocallimastix sp. L2 is monomembraned [10], Benchimol et al. modified the traditional fixation procedure with CaCl2 and, through transmission electron microscopy and frozen sections, found that the inner and outer membranes are very close to each other [9]. This indicated that an appropriate fixation method is necessary for good visualization of the two membranes. In addition, researchers have found that in some regions of the hydrogenosome of Trichomonas foetus (T. foetus), the separation between the two membranes forms a vesicle structure. Under transmission electron microscopy, the electron density of the vesicles was slightly higher than that of the matrix [11]. The number, size, and electron density of the vesicles varied in different microorganisms [11][12][13][14]. These vesicles may be related to the storage of calcium and other cations and change at different metabolic states.
2.2. Function of the Hydrogenosome
In 1973, Lindmark and Muller discovered that the organelles in T. foetus did not possess the typical mitochondrial characteristics of the tricarboxylic acid cycle, electron transport chain, F0–F1 ATPase, and oxidative phosphorylation, but the enzyme system in the particle matrix was capable of completing the anaerobic metabolism of pyruvate. In contrast to mitochondria and peroxisomes, these organelles directly use protons as terminal electron receptors to produce H2; therefore, they proposed a name for this subcellular particle called hydrogenosome in conjunction with previous studies [13]. The hydrogenosome converts pyruvate into various metabolites, such as formate, acetate, H2, and CO2, and also produce ATP through substrate-level phosphorylation for cell growth, acting as an oxygen-independent mitochondrion.
In addition, the hydrogenosome possesses another important function for its survival known as antioxidative stress response. The enzymes in the hydrogenosome, especially pyruvate: ferredoxin oxidoreductase (PFO) and hydrogenase, are highly sensitive to O2 and can be inactivated rapidly under the condition of O2, blocking the substance metabolism in the hydrogenosome. The inhibition of the enzyme by O2 may be due to the formation of reactive oxygen species (ROS). For example, O2 is thought to bind to [Fe] at the far end of the [2Fe]H subcluster, then form an ROS and destroy the [4Fe4S]H subcluster to inactivate the [FeFe] hydrogenase [15]. In most living organisms, iron-dependent superoxide dismutase (SOD) converts ROS to hydrogen peroxide (H2O2), which is subsequently removed by mitochondria usually using the glutathione system and catalase. The mechanism by which the hydrogenosome defends against the damage from ROS is not entirely the same. Although Lindmark et al. reported SOD activity in T. foetus hydrogenosomes [16], there have been no reports of peroxide reductase in the organelles that break down H2O2. Unlike mitochondria, thioredoxin-linked peroxiredoxin antioxidant system is one of the major antioxidant defense mechanisms in trichomonas hydrogenosomes. Three relevant peroxidases in the hydrogenosome of Trichomonas vaginalis (T. vaginalis), thioredoxin reductase (TrxR), thioredoxin (Trx), and Trx-dependent peroxidases (TrxP) jointly play a role in the hydrogenosome to reduce oxidative stress caused by H2O2. Moreover, when T. vaginalis is subjected to oxidative stress, the expression of Trx and TrxP is increased at both transcriptional and translational levels, demonstrating that the parasite is capable of responding to exogenous oxygen stress through changes in both levels [17].
3. Metabolism in the Hydrogenosome
3.1. Carbohydrate Metabolism
Due to the difference in structure and enzyme, the hydrogenosomal metabolism in different microorganisms is not exactly the same. In general, there are many enzymes related to carbohydrate metabolism in the hydrogenosome, including malic enzyme (ME), PFO, [2Fe-2S] ferredoxin protein (Fdx), succinate thiokinase (SCS), adenosine kinase (AK), [Fe] hydrogenase (Hyd), acetate: succinyl CoA-transferase (ASCT), among others. Glucose enters the glycolysis process when it is transferred into the cytoplasm by glucose transporters. Then the formed malate or pyruvate goes into the hydrogenosome; it is metabolized as CO2, formate, and acetate and finally releases energy in the form of ATP. PFO and hydrogenase are two hydrogenosomal enzymes that do not exist in mitochondria. Instead of the pyruvate dehydrogenase enzyme complex, PFO converts the pyruvate produced by glycolysis to acetyl CoA and CO2 (by taking off the hydroxyl group). Then the consequent electron is passed to the middle electronic carrier Fdx, and the reduced Fdx is oxidized by ferredoxin hydrogenase, eventually passing electrons to protons (H+) to form H2. H2 in the hydrogenosome can also be produced in a determined way by hydrogen dehydrogenase; however, the enzyme mainly operates the reverse reaction to converse NAD(P)+ and H2 to H+ and NAD(P)H, which is in an energetically favorable direction. In addition, a speculative way of bifurcating hydrogenase to produce H2 has been put forward and seems important in the coupling of the reduction of H+ to the oxidation of NAD(P)H through the ferredoxin to generate H2 [18]. ATP is formed by substrate-level phosphorylation [19][20] (Figure 3).
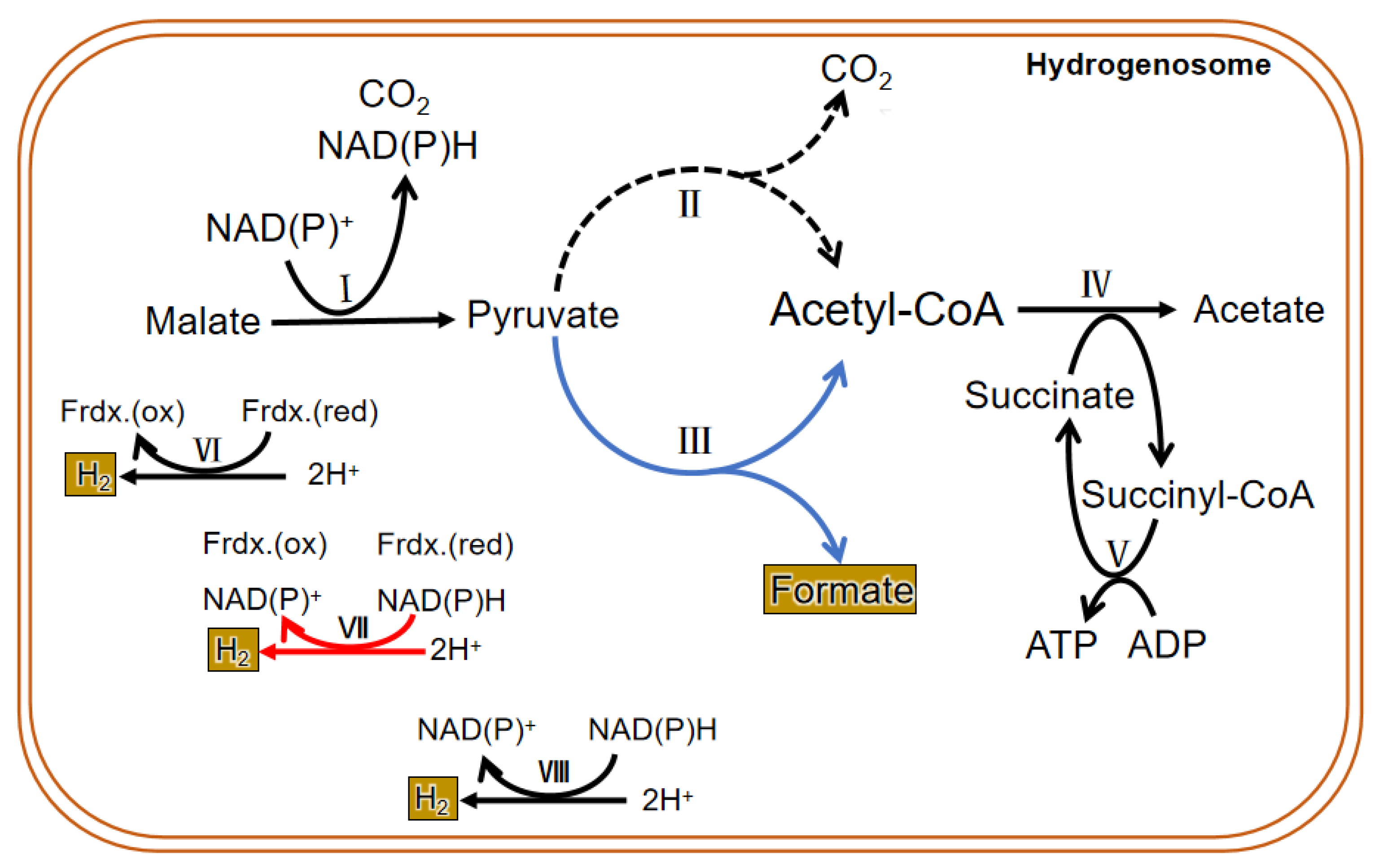
Figure 3. Metabolic process in the hydrogenosome of both trichomonas and anaerobic fungi. The dotted line is the main metabolic pathway of trichomonas hydrogenosomes. The blue line represents the main hydrogenosomal metabolism pathway of anaerobic fungi. The black line is the common metabolic pathway of both of them. The red line shows a suspected H2 generation pathway, and it commonly exits in both. I. malic enzyme; II. pyruvate: ferredoxin oxidoreductase (PFO); III. pyruvate formate lyase (PFL); IV. acetate: succinyl CoA-transferase; V. succinyl-CoA synthetase; VI. ferredoxin hydrogenase; VII. bifurcating hydrogenase; VIII. hydrogen dehydrogenase.
3.2. Amino Acid Metabolism
Carbohydrates are the first source of energy for a living organism. Living organisms will absorb other nutrients, such as amino acids, to sustain its growth when carbohydrates are insufficient. Previous studies have shown that under normal culture conditions, T. vaginalis consumes large amounts of arginine and small amounts of methionine for energy production; and when maltose is deficient, it consumes higher amounts of amino acids, especially arginine, threonine, and leucine [21].
Mukherjee et al. reported the identification and analyses of two glycine cleavage H proteins and a dihydrolipoamide dehydrogenase (L protein) from T. vaginalis, which are pivotal members of the glycine decarboxylase complex, which catalyzes the oxidative decarboxylation and deamination of glycine. They determined their location in the hydrogenosome by immunofluorescence analysis, indicating the important role of the hydrogenosome in amino acid metabolism [22]. The amino acid synthesis and decomposition are associated with the Krebs cycle, glycolytic pathway, and pentose phosphate pathway through conversion to intermediate products (such as pyruvate, acetyl CoA, oxaloacetic acid, acetoacetic acid) in the glucose metabolism in the mitochondria. Similarly, metabolism of amino acids is linked with glycolysis for ATP production in the hydrogenosome. Many enzymes associated with amino acid metabolism and other related metabolic pathways in the protein metabolism at the gene level have been identified [23]. In addition to using immunology at the protein level, or RNA sequencing at the transcriptional level to analyze the differential expression of genes under changing environmental conditions, Huang et al. used LC–FTMS for the first time to analyze the regulation of amino acid metabolism in the hydrogenosome of T. vaginalis and its adaptive response in a glucose restriction environment at the metabolome level. The results showed that amino acid metabolism in the hydrogenosome of T. vaginalis was significantly altered in the case of carbohydrate deficiency. The endergonic metabolism of methionine was less stimulated to reduce ATP consumption, while the branched chain amino acid synthesis related to the glutamate metabolic pathway was markedly enhanced for energy generation [24].
3.3. The Unique Metabolism of the Hydrogenosome in Anaerobic Fungi
The findings on the metabolism of the hydrogenosome are mainly based on the results of experiments conducted in the past several decades using trichomonas as a model. Therefore, the conclusions cannot prove that all hydrogenosomes have the same enzymes and metabolic activities. In fact, the enzyme composition and material metabolism in the hydrogenosome of anaerobic fungi are different from that of other microorganisms (Figure 3). Unlike other anaerobic microorganisms, anaerobic fungi depend on pyruvate: formate lyase (PFL), which exists in both the cytoplasm and the hydrogenosome to hydrolyze pyruvate into isomole formate and acetyl CoA, rather than PFO. Though many studies have suggested that PFO is either absent or of only marginal importance in the anaerobic fungal hydrogenosomal metabolism, both enzymes were identified in all published gut fungal genomes, and a metabolic model proposed and analyzed by Wilken et al. suggested that PFL carries more metabolism flux than PFO in the hydrogenosome, but an energetically favorable route to H2 production still requires the action of PFO [18]. The pyruvate in the cytoplasm is eventually metabolized to ethanol, and the pyruvate that enters in the hydrogenosome is eventually metabolized to acetate. Pyruvate can be directly transported into the hydrogenosome, and also may be formed from the dehydrogenation of malate entered into the hydrogenosome. CO2 and NAD(P)H are also produced during the dehydrogenation of malate, and then hydrogenase reduces 2H+ to H2 with or without the reducing equivalent of NAD(P)H. Acetyl-CoA formed in the hydrolysis of pyruvate first generates acetate and succinyl-CoA under the action of succinyl-CoA transferase. Then succinyl-CoA is converted to succinate by succinyl-CoA synthase, along with the release of CoA and conversion of ADP to ATP, which supplies energy for anaerobic fungi. Therefore, PFL is one of the most important enzymes in the sugar metabolism of anaerobic fungi. Thus, in addition to H2, CO2, acetate, lactate, ethanol, and succinate, the fermentation products of carbohydrates in anaerobic fungi contain formate in equal molar quantities with acetate plus ethanol [25].
So far, there have been no reports on the metabolism of amino acids in the hydrogenosomes of anaerobic fungi.
References
- Gray, M.W. The pre-endosymbiont hypothesis: A new perspective on the origin and evolution of mitochondria. Cold Spring Harb. Perspect. Biol. 2014, 6, a016097.
- Boxma, B.; de Graaf, R.M.; van der Staay, G.W.; van Alen, T.A.; Ricard, G.; Gabaldón, T.; van Hoek, A.H.; Moon-van der Staay, S.Y.; Koopman, W.J.; van Hellemond, J.J.; et al. An anaerobic mitochondrion that produces hydrogen. Nature 2005, 434, 74–79.
- Hackstein, J.H.P.; de Graaf, R.M.; van Hellemond, J.J.; Tielens, A.G.M. Hydrogenosomes of anaerobic ciliates. In Hydrogenosomes and Mitosomes: Mitochondria of Anaerobic Eukaryotes; Tachezy, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 97–112.
- Van der Giezen, M.; Kiel, J.A.; Sjollema, K.A.; Prins, R.A. The hydrogenosomal malic enzyme from the anaerobic fungus Neocallimastix frontalis is targeted to mitochondria of the methylotrophic yeast hansenula polymorpha. Curr. Genet. 1998, 33, 131–135.
- Dacks, J.B.; Dyal, P.L.; Embley, T.M.; van der Giezen, M. Hydrogenosomal succinyl-CoA synthetase from the rumen-dwelling fungus Neocallimastix patriciarum; An energy-producing enzyme of mitochondrial origin. Gene 2006, 373, 75–82.
- van Grinsven, K.W.A.; Rosnowsky, S.; van Weelden, S.W.H.; Pütz, S.; van der Giezen, M.; Martin, W.; van Hellemond, J.J.; Tielens, A.G.M.; Henze, K. Acetate:succinate CoA-transferase in the hydrogenosomes of Trichomonas vaginalis: Identification and characterization. J. Biol. Chem. 2008, 283, 1411–1418.
- Lewis, W.H.; Lind, A.E.; Sendra, K.M.; Onsbring, H.; Williams, T.A.; Esteban, G.F.; Hirt, R.P.; Ettema, T.J.G.; Embley, T.M. Convergent evolution of hydrogenosomes from mitochondria by gene transfer and loss. Mol. Biol. Evol. 2020, 37, 524–539.
- Diniz, J.A.P.; Benchimol, M. Monocercomonas sp.: Cytochemistry and fine structure of freeze-fractured membranes. J. Eukaryot. Microbiol. 1998, 45, 314–322.
- Benchimol, M.; Durand, R.; Almeida, J.C. A double membrane surrounds the hydrogenosomes of the anaerobic fungus Neocallimastix frontalis. FEMS Microbiol. Lett. 1997, 154, 277–282.
- Marvin-Sikkema, F.D.; Lahpor, G.A.; Kraak, M.N.; Gottschal, J.C.; Prins, R.A. Characterization of an anaerobic fungus from llama faeces. J. Gen. Microbiol. 1992, 138, 2235–2241.
- Chapman, A.; Hann, A.C.; Linstead, D.; Lloyd, D. Energy-dispersive X-ray microanalysis of membrane-associated inclusions in hydrogenosomes isolated from Trichomonas vaginalis. J. Gen. Microbiol. 1985, 131, 2933–2939.
- Munn, E.A.; Orpin, C.G. The fine structure of Eadie’s ovals isolated from sheep rumen. J. Gen. Microbiol. 1975, 90, 41–54.
- Lindmark, D.G.; Müller, M. Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J. Biol. Chem. 1973, 248, 7724–7728.
- Bingham, A.S.; Smith, P.R.; Swartz, J.R. Evolution of an hydrogenase with decreased oxygen sensitivity. Int. J. Hydrogrn Energy 2011, 37, 2965–2976.
- Lindmark, D.G.; Müller, M. Superoxide dismutase in the anaerobic flagellates, Tritrichomonas foetus and Monocercomonas sp. J. Biol. Chem. 1974, 249, 4634–4637.
- Coombs, G.H.; Westrop, G.D.; Suchan, P.; Puzova, G.; Hirt, R.P.; Embley, T.M.; Mottram, J.C.; Müller, S. The amitochondriate eukaryote Trichomonas vaginalis contains a divergent thioredoxin-linked peroxiredoxin antioxidant system. J. Biol. Chem. 2004, 279, 5249–5256.
- Biagini, G.A.; Finlay, B.J.; Lloyd, D. Evolution of the hydrogenosome. FEMS Microbiol. Lett. 1997, 155, 133–140.
- Wilken, S.E.; Monk, J.M.; Leggieri, P.A.; Lawson, C.E.; Lankiewicz, T.S.; Seppälä, S.; Daum, C.G.; Jenkins, J.; Lipzen, A.M.; Mondo, S.J.; et al. Experimentally validated reconstruction and analysis of a genome-scale metabolic model of an anaerobic Neocallimastigomycota fungus. mSystems 2021, 6, e00002–e00021.
- Do, P.M.; Angerhofer, A.; Hrdy, I.; Bardonova, L.; Ingram, L.O.; Shanmugam, K.T. Engineering Escherichia coli for fermentative dihydrogen production: Potential role of NADH-ferredoxin oxidoreductase from the hydrogenosome of anaerobic protozoa. Appl. Biochem. Biotechnol. 2009, 153, 21–33.
- Gawryluk, R.M.R.; Kamikawa, R.; Stairs, C.W.; Silberman, J.D.; Brown, M.W.; Roger, A.J. The Earliest Stages of Mitochondrial Adaptation to Low Oxygen Revealed in a Novel Rhizarian. Curr. Biol. 2016, 26, 2729–2738.
- Petrin, D.; Delgaty, K.; Bhatt, R.; Garber, G. Clinical and microbiological aspects of Trichomonas vaginalis. Clin. Microbiol. Rev. 1998, 11, 300–317.
- Mukherjee, M.; Brown, M.T.; McArthur, A.G.; Johnson, P.J. Proteins of the glycine decarboxylase complex in the hydrogenosome of Trichomonas vaginalis. Eukaryot. Cell. 2006, 5, 2062–2071.
- Schneider, R.E.; Brown, M.T.; Shiflett, A.M.; Dyall, S.D.; Hayes, R.D.; Xie, Y.; Loo, J.A.; Johnson, P.J. The Trichomonas vaginalis hydrogenosome proteome is highly reduced relative to mitochondria, yet complex compared with mitosomes. Int. J. Parasitol. 2011, 41, 1421–1434.
- Huang, K.Y.; Ong, S.C.; Wu, C.C.; Hsu, C.W.; Lin, H.C.; Fang, Y.K.; Cheng, W.H.; Huang, P.J.; Chiu, C.H.; Tang, P. Metabolic reprogramming of hydrogenosomal amino acids in Trichomonas vaginalis under glucose restriction. J. Microbiol. Immunol. Infect. 2019, 52, 630–637.
- Boxma, B.; Voncken, F.; Jannink, S.; van Alen, T.; Akhmanova, A.; van Weelden, S.W.; van Hellemond, J.J.; Ricard, G.; Huynen, M.; Tielens, A.G.; et al. The anaerobic chytridiomycete fungus Piromyces sp. E2 produces ethanol via pyruvate:formate lyase and an alcohol dehydrogenase E. Mol. Microbiol. 2004, 51, 1389–1399.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.2K
Revisions:
2 times
(View History)
Update Date:
06 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




