Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jamal Khalife | -- | 1524 | 2022-04-01 10:54:14 | | | |
| 2 | Camila Xu | Meta information modification | 1524 | 2022-04-02 02:42:33 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Khalife, J.; Fréville, A.; , .; Gissot, M.; Pierrot, C. Protein Phosphatases in Apicomplexa. Encyclopedia. Available online: https://encyclopedia.pub/entry/21268 (accessed on 04 March 2026).
Khalife J, Fréville A, , Gissot M, Pierrot C. Protein Phosphatases in Apicomplexa. Encyclopedia. Available at: https://encyclopedia.pub/entry/21268. Accessed March 04, 2026.
Khalife, Jamal, Aline Fréville, , Mathieu Gissot, Christine Pierrot. "Protein Phosphatases in Apicomplexa" Encyclopedia, https://encyclopedia.pub/entry/21268 (accessed March 04, 2026).
Khalife, J., Fréville, A., , ., Gissot, M., & Pierrot, C. (2022, April 01). Protein Phosphatases in Apicomplexa. In Encyclopedia. https://encyclopedia.pub/entry/21268
Khalife, Jamal, et al. "Protein Phosphatases in Apicomplexa." Encyclopedia. Web. 01 April, 2022.
Copy Citation
Apicomplexa correspond to a large and diverse phylum of more than 6000 eukaryotic protozoa that live as obligate parasites in humans and animals. Protein phosphorylation is an ancient-in-origin post-translational modification that is probably universal across phyla. Protein kinases and phosphatases cover between 2% and 4% of a typical eukaryote’s proteome. Unlike protein kinases that catalyze the formation of a covalent bond between a protein substrate and a phosphate group, protein phosphatases catalyze the removal of that phosphate group by hydrolysis.
Apicomplexa
Plasmodium
Toxoplasma
protein phosphatases
1. The Impact of Apicomplexa in Human Health: An Overview
Apicomplexa correspond to a large and diverse phylum of more than 6000 eukaryotic protozoa that live as obligate parasites in humans and animals [1]. Several major human pathogens such as Plasmodium spp. (causing Malaria), Toxoplasma gondii (causing Toxoplasmosis), Cryptosporidium spp. (causing Cryptosporidiosis) and Babesia spp. (causing Babesiosis) belong to this phylum. Apicomplexa are characterized by the presence of a complex of secretory organelles essential to the establishment of host cell infection, namely rhoptries and micronemes [2], and by the presence of a vestigial chloroplast-like organelle called the apicoplast, which is involved in metabolic processes crucial for parasite survival. Cryptosporidium is an exception, being the only Apicomplexa lacking an apicoplast [3][4][5].
Plasmodium, the causative agent of human malaria, is transmitted by Anopheles mosquitoes. It affects about half of the world population and has led to an estimated 229 million cases worldwide, mainly in Africa (93%) but also in Southeast Asia (3.4%) and the Eastern Mediterranean region (2.1%), causing major social, economic and health problems [6]. The clinical manifestations of the disease are linked to the parasite cyclic development in red blood cells and the amplitude of the host immune response. They include fever and flu-like symptoms and can lead to fatal complications such as chronic anemia or cerebral malaria. In 2019, an estimated 409,000 people died from malaria. Despite recent progress linked to local elimination campaigns launched in endemic areas and promises of the RTS,S vaccine recently recommended by the WHO, the capacity of Plasmodium to escape protective host immune responses and the continual emergence of resistance to current treatments and insecticides significantly impede eradication [6].
T. gondii is a widespread opportunistic pathogen affecting between 30% and 60% of the world population. It can infect any nucleated cell in virtually any warm-blooded animal, and its definitive hosts belong to the Felidae family [7][8]. The fetus of newly infected women during pregnancy may be subjected to severe birth defects (e.g., encephalitis and ocular diseases). T. gondii is also an opportunistic pathogen and reactivation of the latent forms in immunocompromised patients can lead to deadly infections [8][9]. Additionally, a growing body of evidence has recently emerged about the possible link between chronic toxoplasmosis and neurological and psychiatric conditions such as schizophrenia or bipolar disorder [10][11][12][13].
Babesia spp. is an intra-erythrocytic parasite transmitted by ticks. Babesia microti and Babesia divergens are the two species predominantly affecting humans, causing flu-like illness and hemolytic anemia. The elderly, immunocompromised or asplenic patients are the most at risk of developing this symptomatic disease [14].
Cryptosporidium is a common pathogen causing moderate-to-severe diarrhea in humans. Two species, Cryptosporidium parvum and Cryptosporidium hominis, are responsible for most cryptosporidiosis cases [15]. The disease can lead to developmental delays and malnourishment, and even death, in children aged 6 to 18 months [16][17]. Cryptosporidiosis is the leading protozoan cause of diarrheal mortality worldwide. Patients with immunocompromised systems are also at risk of developing chronic infection leading to debilitating diarrhea, which may eventually be fatal [18]. Further, cryptosporidiosis may have a causal link with digestive cancers. However, this remains under scrutiny [19].
Apicomplexa are characterized by their diversity, which is tightly linked to the way they evolved to infect specific hosts [20]. Their life cycles are complex, as they require transitions through multiple life stages. These processes can only be achieved through their ability to adjust to changing environments, escape the host defense mechanisms, and undergo massive morphological and metabolic changes [21]. A growing number of studies have recently highlighted the essential role played by phosphatases in the regulation of these processes.
2. Apicomplexan Serine/Threonine and Tyrosine Phosphatome
Protein phosphorylation is an ancient-in-origin post-translational modification that is probably universal across phyla. Protein kinases and phosphatases cover between 2% and 4% of a typical eukaryote’s proteome (reviewed in [22]). Unlike protein kinases that catalyze the formation of a covalent bond between a protein substrate and a phosphate group, protein phosphatases catalyze the removal of that phosphate group by hydrolysis. Another key difference between these two types of enzymes is linked to their structure; while kinases fold according to a single structure model [23], the folding of phosphatases differs according to their catalytic activities. Hence, phosphatases can be grouped into superfamilies [24]. Here, researchers compare the serine/threonine (S/T) and tyrosine (Y) phosphatome of several Apicomplexa of interest (namely, Plasmodium falciparum, Babesia divergens, Toxoplasma gondii and Cryptosporidium parvum) to the S/T/Y human phosphatome. Researchers chose to focus on the enzymes capable of dephosphorylating the S/T/Y residues of proteins, as they represent most of the phosphatome of interest and are well characterized. By comparing the host and parasite phosphatome, researchers aim to highlight the divergences in these enzymes, which are promising sources of potential drug targets.
2.1. Key Differences between the Apicomplexan and Human Phosphatome
When compared to the human S/T/Y phosphatome (140 enzymes), the size of the Apicomplexan S/T/Y phosphatome is much smaller. T. gondii has the biggest phosphatome among the Apicomplexa considered (77 enzymes) and is followed by Plasmodium falciparum (40 enzymes), Cryptosporidium parvum (35 enzymes) and finally Babesia divergens (20 enzymes, see Figure 1). This is in accordance with studies led on the kinomes of these parasites, which found that Apicomplexa parasites presented the smallest kinomes among eukaryotes (from 35 sequences in Babesia bovis up to 135 sequences in Toxoplasma gondii, reviewed in [25]). The reduced number of phosphatases in Apicomplexa as compared to mammalian species is thought to result from the adaptation to a parasitic lifestyle, as parasites can live on nutrients provided by their hosts and thus require less complex metabolic regulation networks (discussed in [26]).
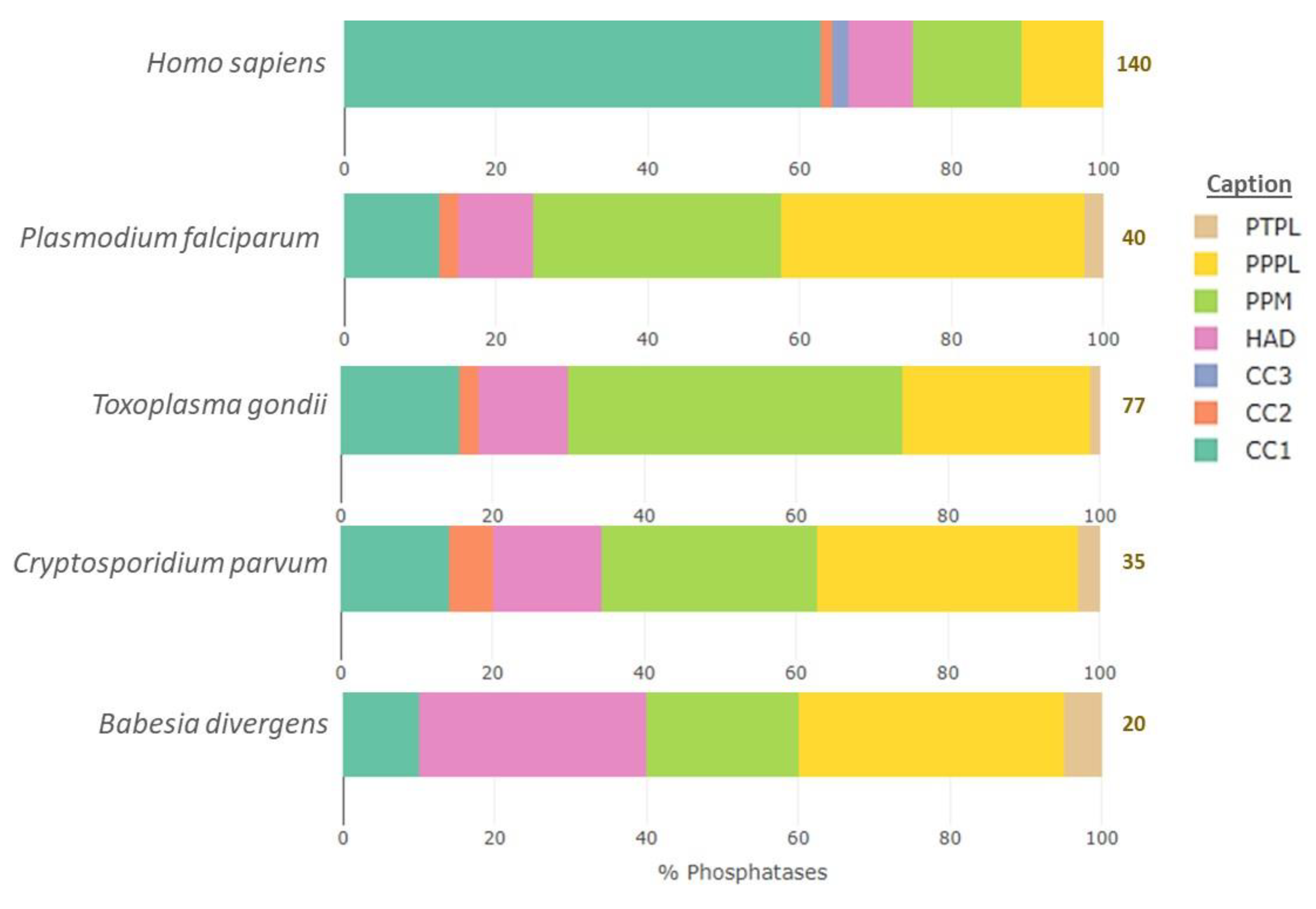 Figure 1. Comparison of the human S/T/Y phosphatome percent composition to those of Plasmodium falciparum, Toxoplasma gondii, Cryptosporidium parvum and Babesia divergens. The total number of phosphatases included for each organism is indicated on the right-hand side of each bar. PTPL, protein tyrosine phosphatase-like; PPPL, phosphoprotein phosphatase like; PPM, protein phosphatases Mn2+ or Mg2+ dependent; HAD, haloacid dehydrogenase; CC1-3, cysteine-based Class I-III. The figure was made using plotly in R version 3.6.3.
Figure 1. Comparison of the human S/T/Y phosphatome percent composition to those of Plasmodium falciparum, Toxoplasma gondii, Cryptosporidium parvum and Babesia divergens. The total number of phosphatases included for each organism is indicated on the right-hand side of each bar. PTPL, protein tyrosine phosphatase-like; PPPL, phosphoprotein phosphatase like; PPM, protein phosphatases Mn2+ or Mg2+ dependent; HAD, haloacid dehydrogenase; CC1-3, cysteine-based Class I-III. The figure was made using plotly in R version 3.6.3.The composition of the protein phosphatome also differs dramatically between the parasites and their intermediate host. The most striking difference is the fact that serine/threonine phosphatases constitute around 80% of the Apicomplexan protein phosphatome but represent only around 30% of the human phosphatome, which contains mostly tyrosine phosphatases (Figure 1).
2.2. General Characteristics of the Apicomplexan Serine/Threonine Phosphatome
According to the classification published by Chen et al. [24], serine/threonine phosphatases can be categorized into three major folds or superfamilies in Apicomplexa, PPPL (phosphoprotein phosphatases-like), PPM/PP2C (protein phosphatase Mn2+ or Mg2+ dependent) and HAD (haloacid dehydrogenase) (see Table 1, Figure 1).
Table 1. Comparison of the human (Hs) S/T/Y phosphatome composition to those of Plasmodium falciparum (Pf), Babesia divergens (Bd), Toxoplasma gondii (Tg) and Cryptosporidium parvum (Cp). Phosphatases are grouped in families following the classification from Chen et al. [24].
| Fold/Superfamily | Family | Substrate | Number in Each Organism | |||||
|---|---|---|---|---|---|---|---|---|
| Hs | Pf | Pb | Bd | Tg | Cp | |||
| PPPL (Phosphoprotein phosphatases-like) |
PPP | pSer/pThr | 13 | 14 | 13 | 6 | 15 | 10 |
| PAP | unknown | 2 | 2 | 2 | 1 | 4 | 2 | |
| PPM (Protein phosphatase Mn2+ or Mg2+-dependent) | PPM | pSer/pThr | 20 | 13 | 13 | 4 | 34 | 10 |
| CC1 (Cysteine-based Class I) |
PTP | pTyr | 37 | 2 | 3 | 1 | 1 | 0 |
| DSP | pTyr, pSer/pThr | 40 | 1 | 1 | 1 | 7 | 5 | |
| DSP (RHOD) | pTyr, pSer/pThr | 11 | 2 | 2 | 0 | 4 | 0 | |
| CC2 (Cysteine-based Class II) |
LMWPTP | pTyr | 1 | 1 | 1 | 0 | 2 | 2 |
| SSU72 | pSer | 1 | 0 | 0 | 0 | 0 | 0 | |
| CC3 (Cysteine-based Class III) |
CDC25 | pTyr, pThr | 3 | 0 | 0 | 0 | 0 | 0 |
| PTPL (Protein tyrosine phosphatase-like) |
PTPLA * | 0 | 1 | 1 | 1 | 1 | 1 | |
| HAD (Haloacid dehydrogenase) |
EYA | pTyr | 4 | 0 | 0 | 0 | 0 | 0 |
| FCP & NIF-like | pSer | 8 | 4 | 5 | 6 | 9 | 5 | |
| Total | 140 | 40 | 41 | 20 | 77 | 35 | ||
(*) Plasmodium PTPLA has been recently re-annotated as DEH (3-hydroxyacyl-CoA dehydratase) and may no longer be considered as a phosphatase.
3. The Function of Protein Phosphatases in Apicomplexa: Where Do We Stand?
In recent years, the massive improvement of reverse genetics and multi-omics technologies has allowed for the development of ambitious projects based on global functional approaches, targeting Apicomplexa of medical importance, such as Plasmodium falciparum and Toxoplasma gondii, as well as the model organism Plasmodium berghei.
Using genome-wide saturation mutagenesis (P. falciparum, [27]), knock-out strategies (P. berghei, [28]) or CRISPR screens (T. gondii, [29]), the contribution of each gene during Plasmodium asexual development or T. gondii development in human fibroblasts was assessed. More than 50% of P. falciparum and 30% of P. berghei phosphatases seem essential for parasite survival in blood cells, highlighting how critical these enzymes are (Figure 2. These studies, along with the characterization of engineered parasite mutant strains, drastically improved the understanding of the critical role played by protein phosphatases over the life cycle of these parasites.
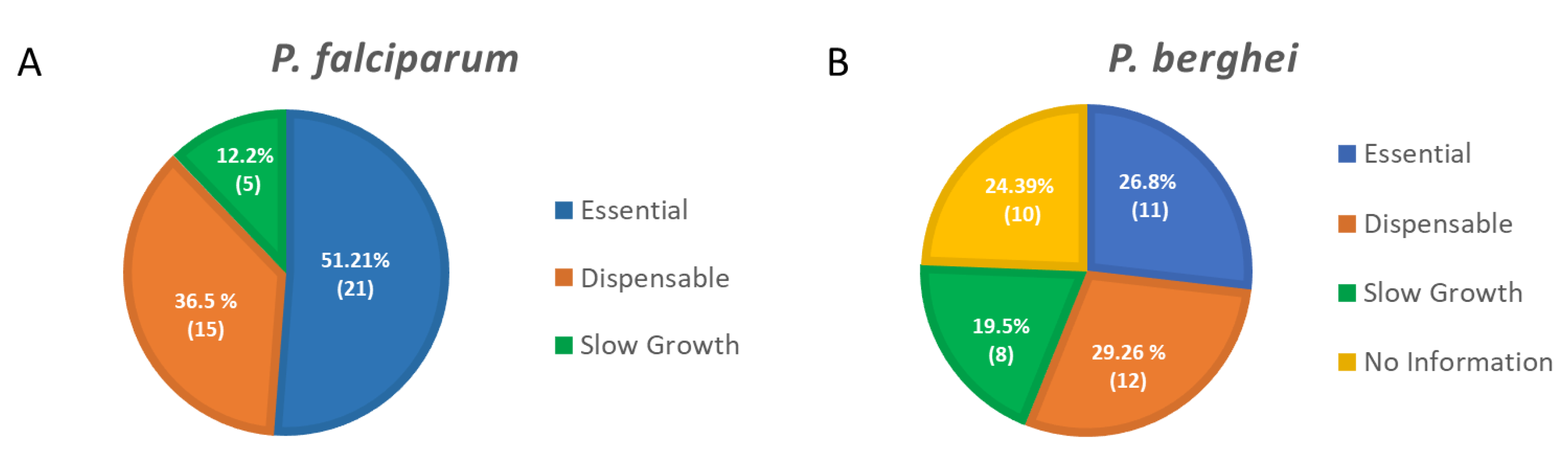 Figure 2. (A). Pie chart of the protein phosphatases’ essentiality in P. falciparum for the parasite blood stage development as determined in the genome-wide saturation mutagenesis [27]. (B). Pie chart of the protein phosphatases’ essentiality in P. berghei for the parasite blood stage development as determined in the PlasmoGEM study [28]. Essential, Dispensable, and Slow Growth represent relative growth rate of 0.1, 1.0, and between 0.1 and 1.0, respectively. The overall percentage and corresponding number of protein phosphatases identified as essential, dispensable or with a slow growth phenotype are represented in the pie charts.
Figure 2. (A). Pie chart of the protein phosphatases’ essentiality in P. falciparum for the parasite blood stage development as determined in the genome-wide saturation mutagenesis [27]. (B). Pie chart of the protein phosphatases’ essentiality in P. berghei for the parasite blood stage development as determined in the PlasmoGEM study [28]. Essential, Dispensable, and Slow Growth represent relative growth rate of 0.1, 1.0, and between 0.1 and 1.0, respectively. The overall percentage and corresponding number of protein phosphatases identified as essential, dispensable or with a slow growth phenotype are represented in the pie charts.References
- Adl, S.M.; Leander, B.S.; Simpson, A.G.B.; Archibald, J.M.; Anderson, O.R.; Bass, D.; Bowser, S.S.; Brugerolle, G.; Farmer, M.A.; Karpov, S.; et al. Diversity, Nomenclature, and Taxonomy of Protists. Syst. Biol. 2007, 56, 684–689.
- Blackman, M.J.; Bannister, L.H. Apical organelles of Apicomplexa: Biology and isolation by subcellular fractionation. Mol. Biochem. Parasitol. 2001, 117, 11–25.
- Fichera, M.E.; Roos, D.S. A plastid organelle as a drug target in apicomplexan parasites. Nature 1997, 390, 407–409.
- McFadden, G.I. Apicoplast. Curr. Biol. 2014, 24, R262–R263.
- Salomaki, E.D.; Kolisko, M. There Is Treasure Everywhere: Reductive Plastid Evolution in Apicomplexa in Light of Their Close Relatives. Biomolecules 2019, 9, 378.
- World Health, O. World Malaria Report 2020: 20 Years of Global Progress and Challenges; World Health Organization: Geneva, Switzerland, 2020.
- Hajj, R.E.; Tawk, L.; Itani, S.; Hamie, M.; Ezzeddine, J.; El Sabban, M.; El Hajj, H. Toxoplasmosis: Current and Emerging Parasite Druggable Targets. Microorganisms 2021, 9, 2531.
- Montoya, J.G.; Liesenfeld, O. Toxoplasmosis. Lancet 2004, 363, 1965–1976.
- Djurković-Djaković, O.; Dupouy-Camet, J.; Van der Giessen, J.; Dubey, J.P. Toxoplasmosis: Overview from a One Health perspective. Food Waterborne Parasitol. 2019, 15, e00054.
- Kezai, A.M.; Lecoeur, C.; Hot, D.; Bounechada, M.; Alouani, M.L.; Marion, S. Association between schizophrenia and Toxoplasma gondii infection in Algeria. Psychiat Res. 2020, 291, 113293.
- Fuglewicz, A.J.; Piotrowski, P.; Stodolak, A. Relationship between toxoplasmosis and schizophrenia: A review. Adv. Clin. Exp. Med. 2017, 26, 1031–1036.
- Fabiani, S.; Pinto, B.; Bonuccelli, U.; Bruschi, F. Neurobiological studies on the relationship between toxoplasmosis and neuropsychiatric diseases. J. Neurol. Sci 2015, 351, 3–8.
- Ngoungou, E.B.; Bhalla, D.; Nzoghe, A.; Dardé, M.-L.; Preux, P.-M. Toxoplasmosis and epilepsy--systematic review and meta analysis. PLoS Negl. Trop. Dis. 2015, 9, e0003525.
- Vannier, E.; Krause, P.J. Human Babesiosis. N. Eng. J. Med. 2012, 366, 2397–2407.
- Feng, Y.; Ryan, U.M.; Xiao, L. Genetic Diversity and Population Structure of Cryptosporidium. Trends Parasitol 2018, 34, 997–1011.
- Platts-Mills, J.A.; Babji, S.; Bodhidatta, L.; Gratz, J.; Haque, R.; Havt, A.; McCormick, B.J.; McGrath, M.; Olortegui, M.P.; Samie, A.; et al. Pathogen-specific burdens of community diarrhoea in developing countries: A multisite birth cohort study (MAL-ED). Lancet Glob. Health 2015, 3, e564–e575.
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222.
- O’Connor, R.M.; Shaffie, R.; Kang, G.; Ward, H.D. Cryptosporidiosis in patients with HIV/AIDS. AIDS 2011, 25, 549–560.
- Sawant, M.; Baydoun, M.; Creusy, C.; Chabé, M.; Viscogliosi, E.; Certad, G.; Benamrouz-Vanneste, S. Cryptosporidium and Colon Cancer: Cause or Consequence? Microorganisms 2020, 8, 1665.
- Van Dooren, G.G.; Striepen, B. The Algal Past and Parasite Present of the Apicoplast. Annu. Rev. Microbiol. 2013, 67, 271–289.
- Gubbels, M.-J.; Coppens, I.; Zarringhalam, K.; Duraisingh, M.T.; Engelberg, K. The Modular Circuitry of Apicomplexan Cell Division Plasticity. Front. Cell Infect. Microbiol. 2021, 11, 670049.
- Moorhead, G.B.G.; De Wever, V.; Templeton, G.; Kerk, D. Evolution of protein phosphatases in plants and animals. Biochem. J. 2009, 417, 401–409.
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934.
- Chen, M.J.; Dixon, J.E.; Manning, G. Genomics and evolution of protein phosphatases. Sci. Signal. 2017, 10, eaag1796.
- Miranda-Saavedra, D.; Gabaldón, T.; Barton, G.J.; Langsley, G.; Doerig, C. The kinomes of apicomplexan parasites. Microbes Infect. 2012, 14, 796–810.
- Miranda-Saavedra, D.; Barton, G.J. Classification and functional annotation of eukaryotic protein kinases. Proteins 2007, 68, 893–914.
- Zhang, M.; Wang, C.; Otto, T.D.; Oberstaller, J.; Liao, X.; Adapa, S.R.; Udenze, K.; Bronner, I.F.; Casandra, D.; Mayho, M.; et al. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 2018, 360, eaap7847.
- Bushell, E.; Gomes, A.R.; Sanderson, T.; Anar, B.; Girling, G.; Herd, C.; Metcalf, T.; Modrzynska, K.; Schwach, F.; Martin, R.E.; et al. Functional Profiling of a Plasmodium Genome Reveals an Abundance of Essential Genes. Cell 2017, 170, 260–272.e8.
- Sidik, S.M.; Huet, D.; Ganesan, S.M.; Huynh, M.H.; Wang, T.; Nasamu, A.S.; Thiru, P.; Saeij, J.P.J.; Carruthers, V.B.; Niles, J.C.; et al. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell 2016, 166, 1423–1435.e12.
More
Information
Subjects:
Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
651
Revisions:
2 times
(View History)
Update Date:
02 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





