You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Robert Y. Tsai | + 3752 word(s) | 3752 | 2022-03-28 05:17:33 | | | |
| 2 | Conner Chen | -40 word(s) | 3712 | 2022-03-31 08:13:41 | | | | |
| 3 | Conner Chen | Meta information modification | 3712 | 2022-03-31 08:14:25 | | | | |
| 4 | Conner Chen | -4 word(s) | 3708 | 2022-04-06 08:21:44 | | | | |
| 5 | Conner Chen | -1 word(s) | 3707 | 2022-04-06 08:22:33 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Tsai, R.Y. The Oral Cancer Prevention. Encyclopedia. Available online: https://encyclopedia.pub/entry/21127 (accessed on 19 December 2025).
Tsai RY. The Oral Cancer Prevention. Encyclopedia. Available at: https://encyclopedia.pub/entry/21127. Accessed December 19, 2025.
Tsai, Robert Y.. "The Oral Cancer Prevention" Encyclopedia, https://encyclopedia.pub/entry/21127 (accessed December 19, 2025).
Tsai, R.Y. (2022, March 28). The Oral Cancer Prevention. In Encyclopedia. https://encyclopedia.pub/entry/21127
Tsai, Robert Y.. "The Oral Cancer Prevention." Encyclopedia. Web. 28 March, 2022.
Copy Citation
Oral cancer is the 18th (out of 36) most common cancer worldwide. Early identification and management of precancerous lesions at high risk of developing cancers is the most effective and economical way to reduce the incidence, mortality, and morbidity of cancers as well as minimizing treatment-related complications, including pain, impaired functions, and disfiguration. Reliable cancer-risk-predictive markers play an important role in enabling evidence-based decision making as well as providing mechanistic insight into the malignant conversion of precancerous lesions.
oral cancer
1. Introduction
Oral cancer is the 18th (out of 36) most common cancer worldwide, with an annual incidence of 377,713 and a mortality of 177,757 in 2020 [1], and is the 8th and 15th most common cancer for males and females in the US, respectively [2]. Its five-year survival rate remains at 66% (American Cancer Society, 2021) (https://www.cancer.org/cacer/oral-cavity-and-oropharyngeal-cancer/detection-diagnosis-staging/survival-rates.html; accessed on 8 March 2022). The etiology of oral squamous cell carcinoma (OSCC) can be categorized into three major groups, including: (1) oral habits associated with tobacco, heavy alcohol, and betel nut chewing; (2) human papilloma virus (HPV) infection; and (3) no known risk factor. In the US, tobacco and heavy alcohol usage remain by far the most common etiological factor, whereas betel nut chewing is more common in Southeast Asia. HPV is a rare cause for OSCC. HPV-negative non-smokers represent a small subset of OSCC patients that are relatively overrepresented by females [3][4][5][6]. Tobacco-related OSCC occurs most commonly on the tongue and the floor of the mouth. It is believed to develop through a premalignant stage of epithelial dysplasia. Dysplastic changes of oral keratinocytes start in the basal and parabasal cell layers, showing hyperchromatism, pleomorphism, increased nuclear-to-cytoplasmic ratio, large and prominent nucleoli, increased mitotic activity, abnormal mitotic figures, and altered epithelial architecture and maturation pattern. Oral epithelial dysplasia is classified either as low-grade, including mild and moderate dysplasia, when cytomorphological changes are confined to the lower half of the epithelium, or as high-grade (severe dysplasia) when changes involve more than half of the epithelial thickness according to the 2017 WHO criteria [7]. This classification was recently challenged for its ability to predict risks, and, as a result, a two-tier grading system was proposed [8]. However, considering the subjectivity in grading and, hence, inter- as well as intra-observer discrepancies, its predictive value requires further validation. This highlights the need for more objective markers for risk prediction of malignant transformation.
2. Risk-Predictive Markers Based on a Longitudinally Followed Study Design
Biomarkers are biological identifiers that can provide crucial information on disease development, diagnosis, and progression. A reliable biomarker needs to demonstrate a high sensitivity and specificity in its power of prediction or differentiation [9]. The terminology used to describe biomarkers serving various clinical purposes can be confusing. Most researched biomarkers fall within three categories: diagnostic, predictive, and prognostic. Diagnostic markers refer to those that differentiate different lesion types or stages without follow-up data (Figure 1A). They are by far the most common type of biomarkers, discovered by comparing cross-sectional samples collected from different patients [10][11][12]. Predictive markers refer to those that indicate the risk of a disease (e.g., cancer) without intervention (Figure 1B). They are discovered by comparing pre-disease/cancer samples with follow-up data indicating their outcome of disease/cancer development. Prognostic markers refer to those that forecast the outcome of a disease/cancer (Figure 1C). They are discovered by comparing disease/cancer samples, after being diagnosed, with follow-up data indicating their outcome with/without intervention. Although diagnostic, predictive, and prognostic markers each serve a particular purpose, occasionally distinction between them can prove difficult. Some diagnostic markers may also play a role in predicting the cancer risk of precancerous lesions or vice versa.
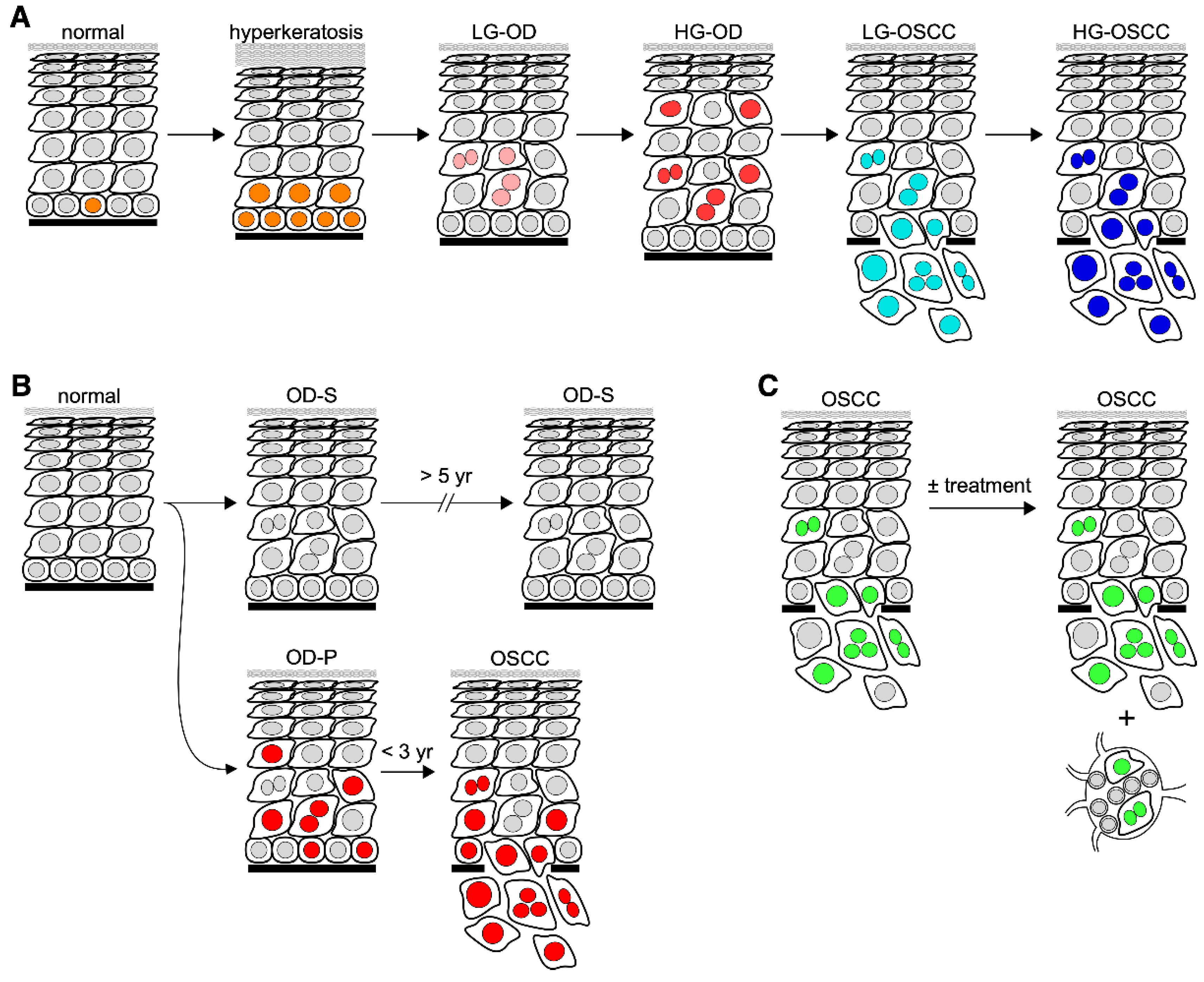
Figure 1. Schematic diagrams of biomarkers in oral carcinogenesis. Diagrams depicting diagnostic markers for differentiating different lesion types (A), predictive markers for assessing the risk of oral premalignant lesions in developing cancer (B), and prognostic markers for predicting the outcome of oral cancer (C). Abbreviations: LG, low-grade; HG, high-grade; OD-P, progressive oral dysplasia; OD-S; static oral dysplasia. Nuclei in (A) are labeled with different color schemes to indicate mitotic cells (orange), low-grade dysplastic cells (pink), high-grade dysplastic cells (red), low-grade malignant cells (light blue), and high-grade malignant cells (dark blue). Nuclei in (B) are labeled in red to indicate cells expressing high-risk markers for malignant progression. Nuclei in (C) are labeled in green to indicate malignant cells.
Current pathological grading of OPLs provides information on the severity of dysplasia that correlates with risk of cancer development to some degree and remains the gold standard in predicting risk of oral cancer in the clinic. High-grade dysplasia has a high (35%) and better predictive value of cancer and, therefore, is recommended for treatment. On the other hand, low-grade dysplasia, which makes up the majority of the OPLs, has a low risk (4–11%) and poor predictive value [13] and is, therefore, not recommended for indiscriminate treatment, given its relatively low transformation rate and most frequent treatment complications, particularly for lesions that are diffuse or large in size. It is, therefore, clinically important to find predictive markers capable of stratifying high-risk vs. low-risk low-grade dysplastic lesions. To date, studies on this topic are quite limited due to the rarity of prospectively collected samples or retrospectively collected pre-progression samples with long-term follow-up data.
3. Predicting the Cancer Risk of OPLs by Quantitative Pathology
The accumulation of genomic instability over time leads to phenotypic changes that can be used to differentiate malignant cells from their normal counterparts. Some of these changes are apparent enough to be detected at the routine (H&E) histopathological level. Under the premise that tumor cells need to acquire sufficient genetic changes to survive and aggressively progress, a subset of at-risk pre-malignant cells may also be phenotypically distinct, and their presence may predict the risk of cancer progression. The emergence of quantitative tissue pathology (QTP) has allowed the profiling of a plethora of microscopic characteristics at the single cell and subcellular level that cannot be gleaned directly by human eyes. These phenotypes could reflect the etiology of cancer and/or the consequence of its underlying pathogenic mechanism [14][15]. Guillaud et al. proposed a promising risk assessment tool, i.e., the nuclear phenotypic score (NPS), for oral cancer progression by building a phenotypic model to recognize nuclear phenotypes, such as nuclear size and shape and DNA amount and distribution, which are significantly discriminative between nuclei of normal, mild, moderate, severe dysplasia, carcinoma in situ, and SCC [16]. Most importantly, based on the best two-group cutoff, NPS was able to separate progressive (high NPS) lesions from non-progressive (low NPS) lesions. Overall, 71% of the high-NPS lesions advanced to cancer within 5 years as comparted to only 22% of the low-NPS lesions, a 10-fold increase in relative risk of progression to cancer based on the NPS level.
4. Cancer-Risk-Predictive Protein Markers for OPLs
Over the last few decades, IHC-based markers have been explored and applied exponentially in many human diseases. Between 1985 and 2006, publications pertaining to IHC markers have increased by over 10,000% [17]. The search terms used to identify studies on IHC-based oral cancer-risk-predictive markers include two core inclusion criteria: (1) primary samples from pre-malignant dysplasia and/or leukoplakia of any grade (low-grade, high-grade, mild, moderate, or severe) collected before they either advanced to cancer (progressive OPLs) or remained OPLs (static OPLs); and (2) samples with longitudinally followed outcomes. Additional terms used for selecting IHC-based studies were risk-predictive, OSCC, predictive biomarkers, pre-malignant oral lesions, immunohistochemistry predictive biomarkers, IHC OSCC risk factors, IHC oral dysplasia, IHC OSCC risk, and/or IHC OSCC prediction. Based on these search criteria, 19 studies were identified, from which 29 IHC-based markers were reported to have the potential of distinguishing progressive vs. static OPLs (Table 1).
4.1. Stem Cell Self-Renewal Factors
Stem cells and cancer cells share many common features, such as self-renewal and undifferentiated potential. Therefore, it should come as no surprise that the most researched protein markers for predicting the development of precancerous lesions into cancers are frequently also implicated in stem cell self-renewal [18]. A study by Zhang et al. reported a higher expression level of β-catenin, a core component of the WNT pathway that is critical for stem cell self-renewal, in OSCC-transformed oral leukoplakia compared to non-transformed oral leukoplakia with a median follow-up of 11.3 years in a univariate analysis (hazard ratio = 4.228, p = 0.001) [19]. The same study also identified cyclooxygenase 2 (COX2), c-Met (also called tyrosine-protein kinase Met or hepatocyte growth factor receptor HGFR), carbonic anhydrase 9 (CA9), Podoplanin (PDPN, a transmembrane glycoprotein associated with lymph node metastasis and poor survival in HNSCC), Ki-67, p16, p53, IMP3 (IMP U3 small nucleolar ribonucleoprotein 3), and c-Jun as potential risk-predictive markers. The WNT pathway also regulates SNAI1 (snail family transcriptional repressor 1) and AXIN2 (axin 2). One study showed that both SNAI1 and AXIN2 were expressed at higher levels in progressive (40.7% for SNAI1 and 26.3% for AXIN2) compared to static leukoplakia (8.7% for SNAI1 and 12.6% for AXIN2), and that both SNAI1 and AXIN2 were independent risk factors for transformation by multivariate analysis [20]. SMAD4 (SMAD family member 4) is a downstream target of the BMP (bone morphogenetic protein) pathway that cross-talks with the WNT pathway [21]. One study reported that low SMAD4 expression in oral leukoplakia is associated with increased malignant transformation and lymphocyte infiltration, suggesting that the combination of low SMAD4 expression and high lymphocyte infiltration may predict the risk of malignant transformation [22]. Notch1 is a cancer stem cell (CSC) marker and a signaling pathway necessary for tissue development and tumor progression. One study reported that oral leukoplakia that progressed to OSCC in five years showed a decrease in nuclear Notch1 expression and an increase in membranous Notch1 expression compared to those that remained static (p = 0.001), and that 38% of patients with membranous expression of Notch1 progressed to OSSC, compared to 13% of those without [23].
The ability to maintain genome integrity throughout DNA replication is an essential feature of self-renewing stem and cancer cells [24]. A 2016 study showed that ATM (ATM serine/threonine kinase) expression was found in 77.8% of progressive dysplasia and 49.4% of static dysplasia, and that yH2AFX expression was observed more in progressive OPLs (55.6%) than in static OPLs (23.5%) [25]. A recent study examined the expression patterns of a stem and cancer cell self-renewal factor, nucleostemin (NS), in human oral dysplastic samples with longitudinally followed outcomes. Nucleostemin is a nucleolar GTP-binding protein that is highly expressed in stem and cancer cells belonging to a novel class of nucleolar GTPases [26][27]. NS plays a crucial role in self-renewal maintenance by promoting the repair of replication-induced DNA damage [28][29][30][31][32][33]. Results revealed that cells with prominent nucleolar NS signals were more abundant in low-grade dysplasia that advanced to OSCC in 2–3 years compared to those remaining static for 7–14 years, suggesting that NS upregulation may be an early event in the progression of low-grade dysplasia to OSCC [34].
SOX2 (SRY-box transcription factor 2) has been implicated in maintaining CSC proliferation in head and neck SCC (HNSCC) [35]. One study showed that the OSCC progression rate in a five-year or longer follow-up period is 44% in patients with positive SOX2 expression and 13% in patients lacking SOX2 expression (p = 0.01) [36]. ALDH1 has also been proposed as a CSC marker for HNSCC. Positive expression of ALDH1 was found at a higher rate (73%) in progressive lesions of oral leukoplakia, either low-grade or high-grade dysplasia, compared to non-progressive lesions (50%) [37]. This study also showed that 58% of oral leukoplakia with positive PDPN progressed to OSCC, compared to only 23% of PDPN negative lesions (p = 0.010). One study reported that oral dysplastic lesions, either low-grade or high-grade, with positive NANOG (Nanog homeobox) expression in the nucleus or cytoplasm showed an increased risk of progression in five years compared to NANOG-negative lesions, and that NANOG expression correlated with the increase in dysplasia grade [38].
4.2. Tumor Suppressors
p53 is a tumor suppressor that plays a master role in determining the outcome of cells (DNA damage repair, cell cycle arrest, or apoptosis) in response to genomic damage. One study indicated that the peak of p53 expression may occur near or during the transition from OPLs to OSCC [39]. A study by Cruz et al. showed that 86% of dysplasias presenting with suprabasal p53 expression (regardless of the grade of dysplasia) progressed to OSCC, as compared to only 22% of dysplasias with negative p53 expression (p = 0.002) [40]. A later study by the same group reported the sensitivity (33%), specificity (83%), positive predictive value (67%), and negative predictive value (56%) of suprabasal p53 expression in predicting the cancer risk of OPLs (i.e., dysplasia and/or leukoplakia) [41]. Loss of another tumor suppressor, p16, was found in both progressive and non-progressive oral leukoplakia, but a significant association with p16 loss was observed only in progressive lesions (p = 0.013) [42].
4.3. Others
MAGE-A (MAGE family member A) proteins are known to be expressed in malignant lesions but not in normal tissue [43]. One study reported a higher MAGE-A expression in oral leukoplakia undergoing malignant transformation in five years compared to those remaining static, with a sensitivity of 85.4% and a specificity of 100% [44]. Another study also showed higher MAGE-A expression in progressive OPLs compared to non-progressive OPLs within a five-year follow-up window, with a positive predictive value of 93% and a negative predictive value of 74.3% [45]. A study by Wu et al. (2018) showed a higher risk of malignant transformation in oral dysplastic lesions with low transglutaminase 3 (TGM3) expression compared to those with high TGM3 expression [46]. One study examined S100A7 overexpression in oral leukoplakia and found 92.3% of progressive OPLs had elevated expression, compared to only 71.8% of non-progressive OPLs (p = 0.014) [47]. A study compared the expression of cortactin and FAK (protein tyrosine kinase 2) in oral dysplasia with a minimum follow up of five years or until malignant transformation and found that those lesions expressing high levels of both proteins displayed the highest incidence of OSCC, followed by those expressing a high level of one of the two proteins and last by those expressing both proteins at low-to-moderate levels [48]. One study utilized a genome-wide expression profile (Bonferroni method) to identify oral leukoplakia with known outcomes collected from a chemoprevention trial [49]. Overexpression of a tyrosine kinase receptor, MET, but not age, treatment arm, or histology, was the only independent predictive factor, showing a hazard ratio of 3.84 (p = 0.003) by multivariate analysis.
Table 1. Cancer-risk-predictive protein markers for OPLs reported by longitudinally designed studies.
| Reference | Biomarker | Conclusions | Functions | Tissue | F/U (Years) | Strength |
|---|---|---|---|---|---|---|
| Zhang et al. [19] | COX-2, c-Met, β-catenin, CA9, PDPN, Ki-67, p16, p53, IMP3, c-Jun | Expression of all markers potentially risk-predictive. Significant differences in positive expression between groups was observed. | Stem Cell Self-Renewal | Oral Leukoplakia (T/N) | 11.3 (median) | 3.04–29.00 (HR) |
| Zhang et al. (2017) [20] | Axin2, Snail | Elevated expression of Snail and Axin2 significantly correlate to risk of malignant transformation. | Stem Cell Self-Renewal | Oral Leukoplakia (T/N) | 10.8 (median) | 4.41, 7.47 (HR) |
| Sakata et al. [22] | SMAD4 | Low expression combined with elevated lymphocyte infiltration indicative of malignant risk. | Stem Cell Self-Renewal | Oral Leukoplakia (T/N) | Unknown | 2.63 (HR) |
| Ding et al. [23] | Notch1 | Expression significantly associated with dysplasia severity and OSCC development. | Stem Cell Self-Renewal | Oral Leukoplakia (T/N) | 6.18 (median) | 3.4 (HR) |
| Crawford et al. [34] | Nucleostemin | NS upregulation may be an early event in malignant transformation of low-grade dysplasia. | Stem Cell Self-Renewal | Oral Dysplasia (P/NP) | 2–3 (NP), 7–14 (P) | p = 0.02–0.05 |
| de Vicente et al. [36] | SOX2 | SOX2 is an independent predictor of cancer risk in OL. | Stem Cell Self-Renewal | Oral Leukoplakia (T/N) | 6.25 (median) | 3.0–5.83 (HR) |
| Habiba et al. [37] | ALDH1, PDPN | Both markers can be used for determining risk of malignant transformation in OL. | Stem Cell Self-Renewal | LG & HG Oral Dysplasia (T/N) | 2.08 (median) | 2.91–3.64 (HR) |
| de Vicente et al. [38] | NANOG | Positive NANOG expression associated with progression to oral cancer-positive expression of both markers demonstrated higher risk. | Stem Cell Self-Renewal | LG & HG Oral Dysplasia (T/N) | 5.08 (median) | 2.01 (HR) |
| Cruz et al. (1998) [40] | p53 | p53 expression pattern has prognostic potential for pre-malignant lesions. | Tumor Suppressor | LG & HG Oral Dysplasia (T/N) | 3 (median) | p = 0.002 |
| Cruz et al. (2002) [41] | P53 | Suprabasal p53 expression is indicative of malignant transformation. | Tumor Suppressor | PMOL (T/N) | 5.0 (mean) | 29–33% Sensitivity,83–100% Specificity |
| Wu et al. [42] | p16 | p16 may predict malignant transformation of OL. | Tumor Suppressor | Oral Leukoplakia (T/N) | Unknown | 3.54 (OR) |
| Baran et al. [45] | MAGE-A | MAGE-A expression can be a reliable predictor of malignant transformation in progressing leukoplakia. | Melanoma Associated Antigen | Oral & Laryngeal Leukoplakia (T/N) | 5 | 96.5% Specificity, 58.2% Sensitivity |
| Ries et al. [44] | MAGE-A | Positive expression in oral leukoplakia is significantly correlated to malignant transformation. | Melanoma Associated Antigen | Oral Leukoplakia (T/N) | 5 | p = 0.0001 |
| Wu et al. [46] | TGM3 | Suggests TGM3 takes part in malignant transformation and may predict progression. | Tumor Suppressor | Oral Leukoplakia (T/N) | 4.75 (T), 7.92 (N) (median) | 5.55 (HR) |
| Kaur et al. [47] | S100A7 | Overexpression demonstrates association with risk of transformation, with cytoplasmic overexpression being most significant. | Cell Cycle & Differentiation | Oral Leukoplakia (T/N) | 3.04 (median) | 2.36 (HR) |
| de Vicente et al. [48] | Cortactin, FAK | Pre-malignant oral lesions with co-expression of both markers demonstrate high risk of OSCC development. | Tumor Progression & Metastasis | Oral dysplasia- leukoplakia, erythroplakia (T/N) | 5 (minimum) | 6.30 (HR) |
| Saintigny et al. [49] | MET | Overexpression in oral leukoplakia was associated with malignant transformation. | Cell Proliferation | Oral Leukoplakia (T/N) | 6.08 (median) | 3.84 (HR) |
| Weber et al. [50] | CD68, CD163 | Elevated CD68 and CD163 significantly associated with malignant transformation. Suggests the value of macrophages as potential predictive markers. | Macrophage Infiltration | Oral dysplasia- mild, moderate, severe (T/N) | 5 (full) | 55.6–72% Sensitivity, 72.7–73.5% Specificity |
| Ries et al. [51] | PD1, PDL1 | Overexpression of both markers may be indicative of cancer risk. | Cell Proliferation | Oral Leukoplakia (T/N) | 5 (minimum) | 50–76.5% Sensitivity, 72.3–93.6% Specificity |
Abbreviations: Follow-Up (F/U), Transformed (T), Non-Transformed (N), Progressing (P), Non-Progressing (NP), hazard ratio (HR), odds ratio (OR).
Finally, a 2020 paper by Weber et al. investigated the possibility of differentiating progressive vs. non-progressive oral leukoplakia based on their tumor immune responses, i.e., macrophage infiltration and polarization [50]. They found that epithelial and subepithelial infiltration of CD68+ and C11+ macrophages were significantly higher in progressive oral leukoplakia compared to non-progressive lesions within the five-year follow-up period. The epithelial density of CD163+ cells was also higher in the progressive than the non-progressive lesions. Another study examined the expression of the immune checkpoint proteins, PD1 (programmed cell death protein 1) and PD-L1 (programmed death-ligand 1) [51]. Oral leukoplakias that transformed in five years showed differences in expression compared to non-transformed leukoplakias, where overexpression of both makers was indicative of malignant transformation. PD1 was significantly overexpressed in both epithelium (p = 0.0001) and sub-epithelium (p = 0.002) in transformed lesions compared to non-transformed lesions. On the other hand, PD-L1 epithelial overexpression nearly reached statistical significance (p = 0.06) and showed a sensitivity of 50% and a specificity of 93.6% in its correlation with progressive lesions.
Despite their wide use in cancer diagnosis and therapeutic practices to provide valuable information on disease progression and prognosis, IHC markers suffer some limitations. IHC staining is subject to a variety of technical variations pertaining to sample acquisition, fixation, processing, preservation, antigen retrieval, and staining procedures. In addition, the significance of the IHC readout is subject to potential interpreter-dependent biases as there is no uniform standard in defining positive vs. negative signals. Finally, most IHC interpretations are qualitative by nature, prone to the subjectivity of the individual analyzing the samples [52].
5. Cancer-Risk-Predictive Genetic Markers for OPLs
Besides the same two core inclusion criteria as described for IHC markers, additional search terms used for selecting studies on genetic markers were risk-predictive, OSCC, predictive biomarkers, pre-malignant oral lesions, genetic predictive biomarkers, genetic OSCC risk factors, genetic OSCC risk, genetic biomarkers oral dysplasia, Loss of heterozygosity (LOH) in oral dysplasia, LOH OSCC prediction, and/or LOH OSCC risk. Based on these search criteria, four studies were identified exploring LOH in similar regions as genetic markers with the potential of distinguishing progressive vs. non-progressive OPLs (Table 2).
Among the many genetic mechanisms that may serve as biomarkers, few have been investigated for their risk-predictive potential. LOH is one that has been frequently studied for its role in malignant transformation of epithelial dysplasia and the development of OSCC. Multiple studies were identified that focused on the risk-predictive potential of LOH. An early 1996 study found that LOH at either 3p and 9p or both was identified in 51% of patients with OPLs and 37% of the 51% patients eventually developed oral cancers, suggesting that LOH in those regions might be early events in tumorigenesis [53]. Another study by Rosin et al. investigated genetic changes between progressive and non-progressive OPLs and determined LOH at regions 3p and 9p to be a necessary feature of progression, as nearly all progressive OPLs harbored this loss. It is worth noting that samples with losses in other regions (4q, 8p, 11q, and 17p) in addition to 3p and 9p also demonstrated a 33-fold increase in cancer risk [54]. One study used a prospectively recruited cohort of low-grade oral dysplasia and confirmed that lesions with LOH in the 3p/9p regions had a 22.6-fold higher risk of malignant transformation compared to lesions with 3p and 9p retention, and that the risk-predictive potential was further increased when combined with LOH at other sites (4q, 17p) [55]. There is also evidence suggesting that a combinatorial approach may increase the cancer-predictive power of LOH by including parameters such as histological changes, chromosomal polysomy, and p53 expression [56]. A later study by Graveland et al. found that lesions with both the 9p LOH and the p53 mutation showed a higher risk of transformation than lesions with 9p LOH alone [57]. Finally, from TCGA molecular profiles of OSCC tumors, OSCC exhibited mutations in tumor suppressor genes at the same loci targeted by LOH, including CDKN2A (cyclin-dependent kinase inhibitor 2A) at 9p21 and TP53. Based on search criteria, none of these reported genetic biomarkers have been validated for their progressive risk by a longitudinal study design.
Table 2. Cancer-risk-predictive genetic markers for OPLs reported by longitudinally designed studies.
| References | Biomarker | Conclusions | Tissue | F/U (Years) | Strength |
|---|---|---|---|---|---|
| Mao et al. [53] | LOH at 3p, 9p | Losses in these regions are frequent early genetic events in OPLs. Cancer developed more quickly in groups with LOH in regions 3p and/or 9p than those without LOH. | Oral Leukoplakia (T/N) | 5.25 (median) | p = 0.039 |
| Rosin et al. [54] | LOH at 3p, 9p, 4q, 8p, 11q, 17p | LOH at 3p and/or 9p exhibit increased risk of cancer development. Risk significantly increased in patients with losses on additional regions. | Hyperplasia, mild and moderate oral dysplasia (P/NP) | 0.5 (minimum) | 3.75, 33.4 (RR) |
| Zhang et al. (2012) [55] | LOH at 3p, 9p, 4q, 17p | LOH at 3p and/or 9p indicates risk for malignant transformation. Risk further increases when combined with LOH at additional sites. | Oral Dysplasia (P/NP) | 3.7 and 3.6 (median) | 22.6 (HR) |
| Graveland et al. [57] | LOH at 9p and p53 mutation | TP53 mutation correlated with losses at 17p and 9p. Losses at 9p significantly associated with risk of transformation. | Oral Leukoplakia (P/NP) | 1.5 (median) | p = 0.014 |
Abbreviations: Follow-Up (F/U), Transformed (T), Non-Transformed (N), Progressing (P), Non-Progressing (NP), hazard ratio (HR), relative risk (RR).
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Neville, B.W.; Damm, D.D.; Allen, C.M.; Bouquot, J.E. Squamous cell carcinoma. In Oral and Maxillofacial Pathology; Saunders Elsevier: St. Louis, MO, USA, 2016; pp. 409–421.
- Chaturvedi, A.K.; Anderson, W.F.; Lortet-Tieulent, J.; Curado, M.P.; Ferlay, J.; Franceschi, S.; Rosenberg, P.S.; Bray, F.; Gillison, M.L. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J. Clin. Oncol. 2013, 31, 4550–4559.
- Koo, K.; Barrowman, R.; McCullough, M.; Iseli, T.; Wiesenfeld, D. Non-smoking non-drinking elderly females: A clinically distinct subgroup of oral squamous cell carcinoma patients. Int. J. Oral Maxillofac. Surg. 2013, 42, 929–933.
- Katzel, J.A.; Merchant, M.; Chaturvedi, A.K.; Silverberg, M.J. Contribution of demographic and behavioral factors on the changing incidence rates of oropharyngeal and oral cavity cancers in northern California. Cancer Epidemiol. Biomark. Prev. 2015, 24, 978–984.
- Conway, D.I.; Brenner, D.R.; McMahon, A.D.; Macpherson, L.M.; Agudo, A.; Ahrens, W.; Bosetti, C.; Brenner, H.; Castellsague, X.; Chen, C.; et al. Estimating and explaining the effect of education and income on head and neck cancer risk: INHANCE consortium pooled analysis of 31 case-control studies from 27 countries. Int. J. Cancer 2015, 136, 1125–1139.
- Reibel, J.; Gale, N.; Hille, J.; Hunt, J.L.; Lingen, M.; Muller, S.; Sloan, P.; Tilakarante, W.M.; Westra, W.H.; Williams, M.D.; et al. Oral potentially malignant disorders and oral epithelial dysplasia. WHO/IARC Classif. Head Neck Tumours 2017, 4, 112–115.
- Odell, E.; Kujan, O.; Warnakulasuriya, S.; Sloan, P. Oral epithelial dysplasia: Recognition, grading and clinical significance. Oral Dis. 2021, 27, 1947–1976.
- Garcia-Gimenez, J.L.; Seco-Cervera, M.; Tollefsbol, T.O.; Roma-Mateo, C.; Peiro-Chova, L.; Lapunzina, P.; Pallardo, F.V. Epigenetic biomarkers: Current strategies and future challenges for their use in the clinical laboratory. Crit. Rev. Clin. Lab. Sci. 2017, 54, 529–550.
- Santosh, A.B.; Jones, T.; Harvey, J. A review on oral cancer biomarkers: Understanding the past and learning from the present. J. Cancer Res. Ther. 2016, 12, 486–492.
- Abati, S.; Bramati, C.; Bondi, S.; Lissoni, A.; Trimarchi, M. Oral cancer and precancer: A narrative review on the relevance of early diagnosis. Int. J. Environ. Res. Public Health 2020, 17, 9160.
- Kitamura, R.; Toyoshima, T.; Tanaka, H.; Kawano, S.; Kiyosue, T.; Matsubara, R.; Goto, Y.; Hirano, M.; Oobu, K.; Nakamura, S. Association of cytokeratin 17 expression with differentiation in oral squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2012, 138, 1299–1310.
- Chi, A.C. Leukoplakia (Leukokeratosis; erythroleukoplakia). In Oral and Maxillofacial Pathology, 4th ed.; Neville, B.W., Damm, D.D., Allen, C.M., Chi, A.C., Eds.; Saunders Elsevier: St. Louis, MO, USA, 2016; pp. 355–363.
- Li, G.; Reinberg, D. Chromatin higher-order structures and gene regulation. Curr. Opin. Genet. Dev. 2011, 21, 175–186.
- Mukhopadhyay, S.; Feldman, M.D.; Abels, E.; Ashfaq, R.; Beltaifa, S.; Cacciabeve, N.G.; Cathro, H.P.; Cheng, L.; Cooper, K.; Dickey, G.E.; et al. Whole slide imaging versus microscopy for primary diagnosis in surgical pathology: A multicenter blinded randomized noninferiority study of 1992 cases (pivotal study). Am. J. Surg. Pathol. 2018, 42, 39–52.
- Guillaud, M.; Zhang, L.; Poh, C.; Rosin, M.P.; MacAulay, C. Potential use of quantitative tissue phenotype to predict malignant risk for oral premalignant lesions. Cancer Res. 2008, 68, 3099–3107.
- Matos, L.L.; Trufelli, D.C.; de Matos, M.G.; da Silva Pinhal, M.A. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark. Insights 2010, 5, 9–20.
- Tsai, R.Y. A molecular view of stem cell and cancer cell self-renewal. Int. J. Biochem. Cell Biol. 2004, 36, 684–694.
- Zhang, X.; Kim, K.Y.; Zheng, Z.; Bazarsad, S.; Kim, J. Nomogram for risk prediction of malignant transformation in oral leukoplakia patients using combined biomarkers. Oral Oncol. 2017, 72, 132–139.
- Zhang, X.; Kim, K.Y.; Zheng, Z.; Kim, H.S.; Cha, I.H.; Yook, J.I. Snail and Axin2 expression predict the malignant transformation of oral leukoplakia. Oral Oncol. 2017, 73, 48–55.
- Du, X.; Li, Q.; Yang, L.; Liu, L.; Cao, Q.; Li, Q. SMAD4 activates Wnt signaling pathway to inhibit granulosa cell apoptosis. Cell Death Dis. 2020, 11, 373.
- Sakata, J.; Yoshida, R.; Matsuoka, Y.; Nagata, M.; Hirosue, A.; Kawahara, K.; Nakamura, T.; Nakamoto, M.; Hirayama, M.; Takahashi, N.; et al. Predictive value of the combination of SMAD4 expression and lymphocyte infiltration in malignant transformation of oral leukoplakia. Cancer Med. 2017, 6, 730–738.
- Ding, X.; Zheng, Y.; Wang, Z.; Zhang, W.; Dong, Y.; Chen, W.; Li, J.; Chu, W.; Zhang, W.; Zhong, Y.; et al. Expression and oncogenic properties of membranous Notch1 in oral leukoplakia and oral squamous cell carcinoma. Oncol. Rep. 2018, 39, 2584–2594.
- Tsai, R.Y. Balancing self-renewal against genome preservation in stem cells: How do they manage to have the cake and eat it too? Cell. Mol. Life Sci. 2016, 73, 1803–1823.
- Zhu, M.; Liu, W.; Shi, L.; Xiao, X.; Wu, W.; Wu, L.; Zhou, Z. Expression of DNA doublestrand repair proteins in oral leukoplakia and the risk of malignant transformation. Oncol. Lett. 2018, 15, 9827–9835.
- Tsai, R.Y.; McKay, R.D. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002, 16, 2991–3003.
- Tsai, R.Y. Turning a new page on nucleostemin and self-renewal. J. Cell Sci. 2014, 127, 3885–3891.
- Meng, L.; Lin, T.; Peng, G.; Hsu, J.K.; Lee, S.; Lin, S.-Y.; Tsai, R.Y. Nucleostemin deletion reveals an essential mechanism that maintains the genomic stability of stem and progenitor cells. Proc. Natl. Acad. Sci. USA 2013, 110, 11415–11420.
- Lin, T.; Ibrahim, W.; Peng, C.-Y.; Finegold, M.J.; Tsai, R.Y. A novel role of nucleostemin in maintaining the genome integrity of dividing hepatocytes during mouse liver development and regeneration. Hepatology 2013, 58, 2176–2187.
- Lin, T.; Meng, L.; Wu, L.J.; Pederson, T.; Tsai, R.Y. Nucleostemin and GNL3L exercise distinct functions in genome protection and ribosome synthesis, respectively. J. Cell Sci. 2014, 127, 2302–2312.
- Lin, T.; Lin, T.C.; McGrail, D.J.; Bhupal, P.K.; Ku, Y.H.; Zhang, W.; Meng, L.; Lin, S.Y.; Peng, G.; Tsai, R.Y.L. Nucleostemin reveals a dichotomous nature of genome maintenance in mammary tumor progression. Oncogene 2019, 38, 3919–3931.
- Wang, J.; McGrail, D.J.; Bhupal, P.K.; Zhang, W.; Lin, K.Y.; Ku, Y.H.; Lin, T.; Wu, H.; Tsai, K.C.; Li, K.; et al. Nucleostemin modulates outcomes of hepatocellular carcinoma via a tumor adaptive mechanism to genomic stress. Mol. Cancer Res. 2020, 18, 723–734.
- Yasumoto, H.; Meng, L.; Lin, T.; Zhu, Q.; Tsai, R.Y. GNL3L inhibits activity of estrogen-related receptor-gamma by competing for coactivator binding. J. Cell Sci. 2007, 120, 2532–2543.
- Crawford, M.; Liu, X.; Cheng, Y.L.; Tsai, R.Y. Nucleostemin upregulation and STAT3 activation as early events in oral epithelial dysplasia progression to squamous cell carcinoma. Neoplasia 2021, 23, 1289–1299.
- Lim, Y.C.; Kang, H.J.; Kim, Y.S.; Choi, E.C. All-trans-retinoic acid inhibits growth of head and neck cancer stem cells by suppression of Wnt/beta-catenin pathway. Eur. J. Cancer 2012, 48, 3310–3318.
- de Vicente, J.C.; Donate-Perez Del Molino, P.; Rodrigo, J.P.; Allonca, E.; Hermida-Prado, F.; Granda-Diaz, R.; Rodriguez Santamarta, T.; Garcia-Pedrero, J.M. SOX2 expression is an independent predictor of oral cancer progression. J. Clin. Med. 2019, 8, 1744.
- Habiba, U.; Hida, K.; Kitamura, T.; Matsuda, A.Y.; Higashino, F.; Ito, Y.M.; Ohiro, Y.; Totsuka, Y.; Shindoh, M. ALDH1 and podoplanin expression patterns predict the risk of malignant transformation in oral leukoplakia. Oncol. Lett. 2017, 13, 321–328.
- de Vicente, J.C.; Rodriguez-Santamarta, T.; Rodrigo, J.P.; Allonca, E.; Vallina, A.; Singhania, A.; Donate-Perez Del Molino, P.; Garcia-Pedrero, J.M. The emerging role of NANOG as an early cancer risk biomarker in patients with oral potentially malignant disorders. J. Clin. Med. 2019, 8, 1376.
- Murti, P.R.; Warnakulasuriya, K.A.; Johnson, N.W.; Bhonsle, R.B.; Gupta, P.C.; Daftary, D.K.; Mehta, F.S. p53 expression in oral precancer as a marker for malignant potential. J. Oral Pathol. Med. 1998, 27, 191–196.
- Cruz, I.B.; Snijders, P.J.; Meijer, C.J.; Braakhuis, B.J.; Snow, G.B.; Walboomers, J.M.; van der Waal, I. p53 expression above the basal cell layer in oral mucosa is an early event of malignant transformation and has predictive value for developing oral squamous cell carcinoma. J. Pathol. 1998, 184, 360–368.
- Cruz, I.; Napier, S.S.; van der Waal, I.; Snijders, P.J.; Walboomers, J.M.; Lamey, P.J.; Cowan, C.G.; Gregg, T.A.; Maxwell, P.; Meijer, C.J. Suprabasal p53 immunoexpression is strongly associated with high grade dysplasia and risk for malignant transformation in potentially malignant oral lesions from Northern Ireland. J. Clin. Pathol. 2002, 55, 98–104.
- Wu, W.; Wang, Z.; Zhou, Z. Role of the human papillomavirus in malignant transformation of oral leukoplakia distinct from oropharyngeal squamous cell carcinoma: A study of 76 patients with internal-control specimens. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019, 128, 273–279.
- Xiao, J.; Chen, H.S. Biological functions of melanoma-associated antigens. World J. Gastroenterol. 2004, 10, 1849–1853.
- Ries, J.; Agaimy, A.; Vairaktaris, E.; Gorecki, P.; Neukam, F.W.; Strassburg, L.H.; Nkenke, E. Detection of MAGE-A expression predicts malignant transformation of oral leukoplakia. Cancer Investig. 2012, 30, 495–502.
- Baran, C.A.; Agaimy, A.; Wehrhan, F.; Weber, M.; Hille, V.; Brunner, K.; Wickenhauser, C.; Siebolts, U.; Nkenke, E.; Kesting, M.; et al. MAGE-A expression in oral and laryngeal leukoplakia predicts malignant transformation. Mod. Pathol. 2019, 32, 1068–1081.
- Wu, X.; Wang, R.; Jiao, J.; Li, S.; Yu, J.; Yin, Z.; Zhou, L.; Gong, Z. Transglutaminase 3 contributes to malignant transformation of oral leukoplakia to cancer. Int. J. Biochem. Cell Biol. 2018, 104, 34–42.
- Kaur, J.; Matta, A.; Kak, I.; Srivastava, G.; Assi, J.; Leong, I.; Witterick, I.; Colgan, T.J.; Macmillan, C.; Siu, K.W.; et al. S100A7 overexpression is a predictive marker for high risk of malignant transformation in oral dysplasia. Int. J. Cancer 2014, 134, 1379–1388.
- de Vicente, J.C.; Rodrigo, J.P.; Rodriguez-Santamarta, T.; Lequerica-Fernandez, P.; Allonca, E.; Garcia-Pedrero, J.M. Cortactin and focal adhesion kinase as predictors of cancer risk in patients with premalignant oral epithelial lesions. Oral Oncol. 2012, 48, 641–646.
- Saintigny, P.; William, W.N., Jr.; Foy, J.P.; Papadimitrakopoulou, V.; Lang, W.; Zhang, L.; Fan, Y.H.; Feng, L.; Kim, E.S.; El-Naggar, A.K.; et al. Met receptor tyrosine kinase and chemoprevention of oral cancer. J. Natl. Cancer Inst. 2018, 110, 250–257.
- Weber, M.; Wehrhan, F.; Baran, C.; Agaimy, A.; Buttner-Herold, M.; Ozturk, H.; Neubauer, K.; Wickenhauser, C.; Kesting, M.; Ries, J. Malignant transformation of oral leukoplakia is associated with macrophage polarization. J. Transl. Med. 2020, 18, 11.
- Ries, J.; Agaimy, A.; Wehrhan, F.; Baran, C.; Bolze, S.; Danzer, E.; Frey, S.; Jantsch, J.; Most, T.; Buttner-Herold, M.; et al. Importance of the PD-1/PD-L1 axis for malignant transformation and risk assessment of oral leukoplakia. Biomedicines 2021, 9, 194.
- Seidal, T.; Balaton, A.J.; Battifora, H. Interpretation and quantification of immunostains. Am. J. Surg. Pathol. 2001, 25, 1204–1207.
- Mao, L.; Lee, J.S.; Fan, Y.H.; Ro, J.Y.; Batsakis, J.G.; Lippman, S.; Hittelman, W.; Hong, W.K. Frequent microsatellite alterations at chromosomes 9p21 and 3p14 in oral premalignant lesions and their value in cancer risk assessment. Nat. Med. 1996, 2, 682–685.
- Rosin, M.P.; Cheng, X.; Poh, C.; Lam, W.L.; Huang, Y.; Lovas, J.; Berean, K.; Epstein, J.B.; Priddy, R.; Le, N.D.; et al. Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clin. Cancer Res. 2000, 6, 357–362.
- Zhang, L.; Poh, C.F.; Williams, M.; Laronde, D.M.; Berean, K.; Gardner, P.J.; Jiang, H.; Wu, L.; Lee, J.J.; Rosin, M.P. Loss of heterozygosity (LOH) profiles—Validated risk predictors for progression to oral cancer. Cancer Prev. Res. 2012, 5, 1081–1089.
- Lee, J.J.; Hong, W.K.; Hittelman, W.N.; Mao, L.; Lotan, R.; Shin, D.M.; Benner, S.E.; Xu, X.C.; Lee, J.S.; Papadimitrakopoulou, V.M.; et al. Predicting cancer development in oral leukoplakia: Ten years of translational research. Clin. Cancer Res. 2000, 6, 1702–1710.
- Graveland, A.P.; Bremmer, J.F.; de Maaker, M.; Brink, A.; Cobussen, P.; Zwart, M.; Braakhuis, B.J.; Bloemena, E.; van der Waal, I.; Leemans, C.R.; et al. Molecular screening of oral precancer. Oral Oncol. 2013, 49, 1129–1135.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
5 times
(View History)
Update Date:
06 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




