You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Blanca Lorente-Torres | + 1991 word(s) | 1991 | 2022-03-15 02:35:52 | | | |
| 2 | Dean Liu | Meta information modification | 1991 | 2022-03-25 04:48:45 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lorente-Torres, B. MicroRNAs in the Treatments against Human Bacterial Pathogens. Encyclopedia. Available online: https://encyclopedia.pub/entry/20969 (accessed on 02 January 2026).
Lorente-Torres B. MicroRNAs in the Treatments against Human Bacterial Pathogens. Encyclopedia. Available at: https://encyclopedia.pub/entry/20969. Accessed January 02, 2026.
Lorente-Torres, Blanca. "MicroRNAs in the Treatments against Human Bacterial Pathogens" Encyclopedia, https://encyclopedia.pub/entry/20969 (accessed January 02, 2026).
Lorente-Torres, B. (2022, March 24). MicroRNAs in the Treatments against Human Bacterial Pathogens. In Encyclopedia. https://encyclopedia.pub/entry/20969
Lorente-Torres, Blanca. "MicroRNAs in the Treatments against Human Bacterial Pathogens." Encyclopedia. Web. 24 March, 2022.
Copy Citation
The development of RNA-based anti-infectives has gained interest with the successful application of mRNA-based vaccines. Small RNAs are molecules of RNA of <200 nucleotides in length that may control the expression of specific genes. Small RNAs include small interference RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), or microRNAs (miRNAs).
miRNAs
pathogen
bacteria
infection
1. Introduction
Small RNAs are non-coding molecules of RNA of less than 200 nucleotides in length and with important roles in transcriptional regulation. There are different small RNAs, such as small interference RNAs (siRNAs), piwi-interacting RNAs (piRNAs), and micro RNAs (miRNAs). MicroRNAs were identified in the early 1990s [1], and their function as transcriptional regulators was gradually elucidated [2]. MiRNAs are typically around 18–25 nucleotides long non-coding molecules that act as transcriptional regulators by targeting specific messenger RNAs (mRNAs) for their destruction to achieve gene silencing [3][4]. MicroRNA biogenesis is an important process that finalizes with the RNA-Induced Silencing Complex (RISC) formation, which localizes and binds miRNA with its target mRNA. This results in the degradation of the targeted mRNA(s) and a subsequent reduction in the expression of the affected gene(s) [5][6][7]. However, the silencing of a transcriptional repressor may result in downstream gene upregulation. Therefore, some miRNAs can also trigger the expression of specific genes [8].
Most mammalian mRNAs possess conserved targets for miRNAs [9]. The perfect match between a miRNA and its target (3’UTR region of the mRNA) results in mRNA cleavage, eventually leading to gene silencing. However, there is also a possibility of a non-perfect match between a miRNA and the 5’UTR region of a gene [10]. The complementarity degree between a miRNA and its mRNA target dictates its level of degradation or silencing [7][11]. In addition, some post-transcriptional alterations could change the processing of miRNAs by the DROSHA/DICER complex and their loading onto Argonaute (AGO) proteins, an essential component of RISC. These changes in miRNA maturation may alter the miRNA-mediated regulation of gene expression and could be different depending on the type of cell or their microenvironment [7].
Despite their highly complex and understudied roles, it is now becoming clear that miRNAs are essential molecules that regulate multiple molecular pathways in humans and other organisms. There are >2500 human miRNAs annotated in public repositories, and >3000 miRNAs have been additionally identified in specific cell types. These >5500 miRNAs have >45,000 gene targets, representing more than 60% of all human protein-coding genes [4][9][12][13][14].
2. Role of miRNAs in the Control of the Infected Host Cell
2.1. Mycobacterium tuberculosis
Autophagy plays an important role during intracellular infection caused by M. tuberculosis, and miRNAs regulate this molecular pathway. The expression of miR-155 and miR-17-5p reduces the intracellular colonisation of M. tuberculosis by modulating different metabolic routes that result in autophagy activation in macrophages [15][16][17]. The upregulation of miR-155 results in the activation of autophagy and a concomitant mycobacterial clearance. In particular, miR-155 binds to the 3’UTR region of the Rheb gene, promoting phagosome maturation, binding to lysosomes, and subsequent mycobacterial elimination [18].
In addition, M. tuberculosis can also control the expression of different miRNAs to reduce autophagy. For instance, miR-27 is upregulated during M. tuberculosis infection and downregulates calcium-associated autophagy [19]. The target of miR-27 is the Ca2+ transported CAC-NA2D3, which is located at the endoplasmic reticulum (ER) and whose downregulation inhibits autophagosome formation [19]. Moreover, M. tuberculosis host cell infection induces the expression of miR-1958, which binds to the 3’UTR region of Atg5, whose silencing results in the inhibition of the autophagic flux [20]. Finally, miR-18a facilitates M. tuberculosis infection by silencing the ataxia–telangiectasia-mutated (ATM) gene, which decreases LC3-II levels in infected cells and stops the xenophagy process [21].
On the other hand, miR-33 is overexpressed during M. tuberculosis infection, and it targets different host cell genes involved in cholesterol transport and fatty acid oxidation, including ABCA1, CROT, CPT1, HADHB and PRKAA1. This, in turn, activates the lipid catabolism in the infected host cells, which facilitates bacterial colonisation because of the highly lipid-dependent metabolism of M. tuberculosis [22]. Thus, an anti-miR-33 may promote phagosome maturation and bacterial clearance by stopping the lipid metabolism of the infected host cell.
2.2. Adherent–Invasive Escherichia coli
Some microRNAs are also relevant in the Adherent–Invasive Escherichia coli (AIEC) colonisation of intestinal mucosa in Crohn’s disease patients [23]. In this context, exosomes carrying miR-30c and miR-130a are released into non-infected cells to silence the expression of ATG5 and ATG16L1. The resulting inhibition of the autophagic flux facilitates the intracellular replication of AIEC [24].
2.3. Legionella pneumophila
Interestingly, Legionella pneumophila may control the expression of 85 different miRNAs during infection. In particular, the upregulation of three miRNAs (miR-125b, miR-221, and miR-579) in a cooperative manner leads to the downregulation of the RNA receptor DDX58/RIG-I, the tumour suppressor TP53, the antibacterial LGALS8 and the MX dynamin-like GTPase 1 (MX1), which altogether enhance the intracellular replication of the pathogen [25]. The repressive effects of miR-125b and miR-221 on MX1 and miR-579 on LGALS8 are particularly significant. These genes form a newly discovered cellular immune response pathway against L. pneumophila whose overexpression results in bacterial clearance [25].
2.4. Chlamydia trachomatis
C. trachomatis maintains mitochondrial ATP production during infection through the upregulation of miR-30c-5p. This miRNA downregulates p53, which in turn leads to the downregulation of Drp1, a mitochondrial fission regulator [26]. Many other intracellular pathogens have very tight interactions with mitochondria during host cell infection, which opens the way to interventions aimed at disrupting bacterial proliferation [27].
2.5. Shigella flexneri and Salmonella enterica Serovar Typhimurium
High-throughput screenings have quickly identified novel microRNAs involved in other bacterial infections. One such study has uncovered three important miRNAs expressed during Shigella flexneri infection: miR-3668, miR-4732-5p and miR-6073. These miRNAs constrain the infection caused by S. flexneri by inhibiting the expression of N-WASP, which in turn restricts bacterial actin-based motility, stops cell-to-cell spread, and attenuates intracellular infection [13]. In contrast, the expression of miR-29b-2-5p promotes the production of filopodia in host cells by targeting Unc-5 Netrin Receptor C (UNC5C), which enhances bacterial uptake [28].
Despite the similarities between Shigella flexneri and Salmonella enterica serovar Typhimurium, the control of the expression of specific miRNAs elicited by both pathogens completely differs [13]. In particular, miR-let-7i-3p targets the host RGS2 protein and modulates vacuolar trafficking during S. Typhimurium infection, inhibiting its pathogenesis [13].
In addition, the miR-15 family of miRNAs is very important in the pathogenesis of S. Typhimurium, as they are downregulated during specific stages of the infection to allow bacterial spreading [29]. In particular, the miR-15 family arrests the cell cycle of infected cells through the inhibition of the transcription factor E2F1 and derepression of cyclin D1 [29][30].
2.6. Burkholderia pseudomallei
It is also important to consider that the balance between pro-infection and anti-infection miRNAs may be determinant in the fate of intracellular bacterial pathogens. For example, Burkholderia pseudomallei downregulates the expression of miR-30b/30c, which results in the upregulation of Rab32. This GTPase promotes the fusion between phagosomes and lysosomes by releasing hydrolases that limit the intracellular growth of B. pseudomallei [31]. However, the expression of miR-3473 is triggered during B. pseudomallei infection of macrophages, which is mediated by the overexpression of TNF receptor-associated factor 3 (TRAF3) and subsequent TNF-α release, favouring bacterial replication [32].
2.7. Listeria monocytogenes
Listeriosis triggers the upregulation of miR-146a, and the silencing of this miRNA reduces the pathogen’s ability to colonise macrophages intracellularly [33]. At the same time, miR-21 is also activated during infection and controls the polarisation of macrophages [34], which eventually leads to a reduction in the intracellular survival of L. monocytogenes [35]. In contrast, miR-26a controls the infection of L. monocytogenes by targeting the Ephrin receptor tyrosine kinase 2 (EphA2) to inhibit the internalization or the phagosomal escape of the pathogen [36]. Intriguingly, EphA2 is also an invasion receptor for C. trachomatis or S. aureus [37][38].
3. Novel Antimicrobial Treatments Based on miRNA-Based Technology
RNA-based technology is becoming a feasible strategy to control bacterial infections [39][40][41][42][43]. This new approach has been successfully tested against pulmonary tuberculosis by employing siRNAs targeting tfgb1 [44]. However, there are now many other opportunities to develop antimicrobial strategies based on other small RNAs, such as many of the miRNAs listed in the previous section.
In addition, the expression of anti-miRNAs targeting specific miRNAs that facilitate bacterial infection may delay or disrupt the pathogen’s host colonisation. Anti-miRNAs are artificially produced single-stranded RNAs that are complementary to target miRNAs and block their functioning [39]. This strategy has been previously applied in the context of viral infections [45][46]. For example, miravirsen is an anti-miRNA that targets miR-122, an essential miRNA during hepatitis C virus (HCV) infection. Results from a phase II clinical trial indicate that miravirsen can reduce the viral load in a dose-dependent manner [47][48]. The same strategy could be potentially applied to silence miRNAs that are essential for the replication of bacterial intracellular pathogens.
However, there are some important challenges in the clinical application of miRNA as anti-infectives. The most significant handicap of miRNA therapies is their off-target effects. This could be due to miRNA interactions in a non-specific manner with partially complementary mRNAs [10], leading to important side effects in the host [41].
Moreover, the delivery of miRNAs to infected cells could be complicated by the presence of RNAses that can quickly degrade them. This could be partially solved by improving the delivery method of microRNAs to reach specific targets at the cellular or even subcellular levels. This problem has been approached from different perspectives, including the use of nanoparticles, viral delivery systems, high-density lipoproteins, liposomes, or exosomes [41][42], which can facilitate their delivery to host cells [49][50].
Currently, lipid nanoparticles are the leading non-viral delivery systems in the clinical setting [51]. Liposomes are a group of lipid particles that are extensively used to guide RNA-based therapies [52]. However, the main disadvantage of liposomes is the difficulty in functionalising their lipid bilayer [53]. Thus, naturally produced extracellular vesicles are now considered an exciting alternative to improve miRNA delivery (Figure 1).
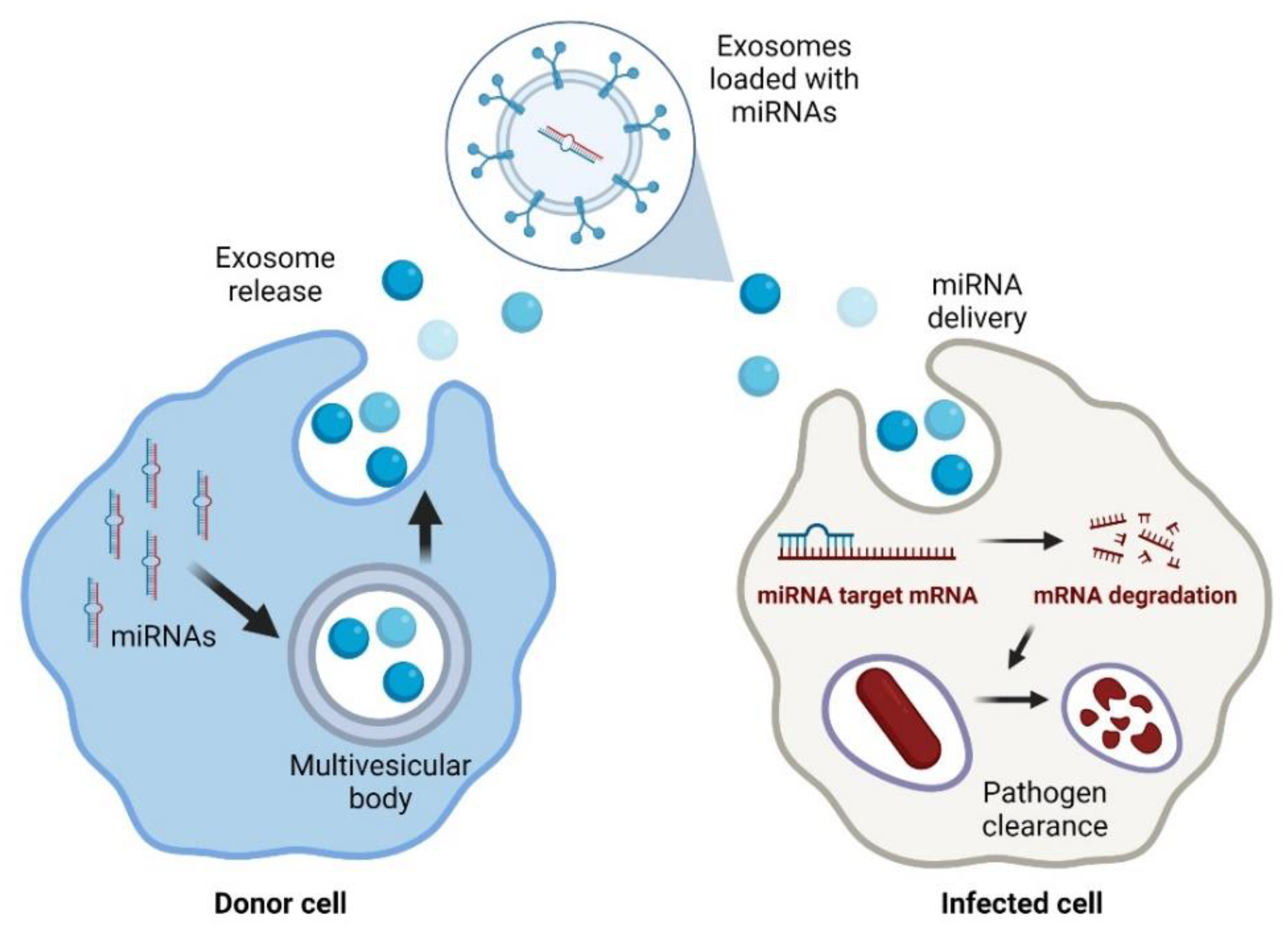
Figure 1. Exosomal delivery of antimicrobial miRNAs to infected cells. Created with BioRender.com (accessed on 1 February 2022).
In particular, exosomes are an up-and-coming solution since they are not toxic and have low antigenicity because they are part of the natural intercellular communication pathways [54]. Exosomes are part of the vesicles generated within the endosomal system and then secreted to the extracellular milieu with essential roles in cell-to-cell communication. Exosomes may efficiently protect the miRNA molecules from degradation by nucleases. Because of this, their use for the delivery of treatments based on nucleic acids is rapidly increasing [55]. In addition, exosomes have advantages over other delivery strategies, such as those based on adenoviruses that may be neutralised by antibodies [54].
Similar to other RNA-delivery systems, exosomes must be modified to target infected cells [26][27]. Cancer research has provided different molecular strategies to increase the specificity of exosomal RNA delivery [56]. The main interactions between exosomes and target cells are mainly based on tetraspanins, integrins, lipids, lectins, heparan sulphate proteoglycans, and extracellular matrix elements [55].
Interestingly, the isolation of exosomes naturally produced by specific cells increased their fusion with the same parental cells. Thus, isolating exosomes derived from tumour cells and loaded with anti-cancer drugs resulted in well-targeted drug delivery [56]. Moreover, changes in the transmembrane proteins present on the surface of exosomes result in a better adhesion to targeting cells [57]. In addition, the rationale design of exosomes with different membrane modifications also showed promising results in vitro and in vivo in cancer therapies [58]. The use of carbonate apatite or glycan polymers has improved the target cell selectivity by increasing the delivery from endosomes to the cytosol of target cells. Thus, the use of carbonate apatite increased the delivery into the liver, and poly-L-lysine-lactose increased the uptake for hepatocytes [59].
Similar approaches could be used to target bacterial-infected cells. However, the development of exosomes as an efficient RNA-delivery system to treat bacterial infections is still in its early stages [60]. During bacterial infection, both the pathogen and eukaryotic cells can produce exosomes that stimulate the immune system or facilitate bacterial infection [61][62]. In addition, exosomes derived from cells primed with bacterial lipopolysaccharide (LPS) could target specific macrophage populations more efficiently and elicit their activation [63]. This strategy may increase the specificity of exosomal-delivery of small RNAs and lower the minimal inhibitory concentration of exosomes required to block host cell infection caused by intracellular pathogens [64][65][66]. Nonetheless, more research is needed to develop an efficient, scalable, easy to produce, stable and specific small RNA delivery system that could be used in the context of bacterial infection.
References
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862.
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20.
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655.
- Kim, J.K.; Kim, T.S.; Basu, J.; Jo, E.K. MicroRNA in innate immunity and autophagy during mycobacterial infection. Cell. Microbiol. 2017, 19, e12687.
- Eulalio, A.; Schulte, L.N.; Voge, J. The mammalian microRNA response to bacterial infections. RNA Biol. 2012, 9, 742–750.
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108.
- de Sousa, M.C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ action through miRNA editing. Int. J. Mol. Sci. 2019, 20, 6249.
- Orang, A.V.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int. J. Genom. 2014, 2014, 970607.
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105.
- Wang, Z.; Rao, D.D.; Senzer, N.; Nemunaitis, J. RNA interference and cancer therapy. Pharm. Res. 2011, 28, 2983–2995.
- Trobaugh, D.W.; Klimstra, W.B. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 2017, 23, 80–93.
- Asl, E.R.; Amini, M.; Najafi, S.; Mansoori, B.; Mokhtarzadeh, A.; Mohammadi, A.; Lotfinejad, P.; Bagheri, M.; Shirjang, S.; Lotfi, Z.; et al. Interplay between MAPK/ERK signaling pathway and MicroRNAs: A crucial mechanism regulating cancer cell metabolism and tumor progression. Life Sci. 2021, 119499.
- Aguilar, C.; Cruz, A.R.; Rodrigues Lopes, I.; Maudet, C.; Sunkavalli, U.; Silva, R.J.; Sharan, M.; Lisowski, C.; Zaldívar-López, S.; Garrido, J.J.; et al. Functional screenings reveal different requirements for host microRNAs in Salmonella and Shigella infection. Nat. Microbiol. 2020, 5, 192–205.
- Londina, E.; Lohera, P.; Telonis, A.G.; Quann, K.; Clark, P.; Jinga, Y.; Hatzimichael, E.; Kirino, Y.; Honda, S.; Lally, M.; et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc. Natl. Acad. Sci. USA 2015, 112, E1106–E1115.
- Silwal, P.; Kim, Y.S.; Basu, J.; Jo, E.K. The roles of microRNAs in regulation of autophagy during bacterial infection. Semin. Cell Dev. Biol. 2020, 101, 51–58.
- Kumar, R.; Sahu, S.K.; Kumar, M.; Jana, K.; Gupta, P.; Gupta, U.D.; Kundu, M.; Basu, J. MicroRNA 17-5p regulates autophagy in Mycobacterium tuberculosis-infected macrophages by targeting Mcl-1 and STAT3. Cell. Microbiol. 2016, 18, 679–691.
- Rothchild, A.C.; Sissons, J.R.; Shafiani, S.; Plaisier, C.; Min, D.; Mai, D.; Gilchrist, M.; Peschon, J.; Larson, R.P.; Bergthaler, A.; et al. MiR-155-regulated molecular network orchestrates cell fate in the innate and adaptive immune response to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2016, 113, E6172–E6181.
- Wang, J.; Yang, K.; Zhou, L.; Wu, M.; Wu, Y.; Zhu, M.; Lai, X.M.; Chen, T.; Feng, L.; Li, M.; et al. MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathog. 2013, 9, e1003697.
- Liu, F.; Chen, J.; Wang, P.; Li, H.; Zhou, Y.; Liu, H.; Liu, Z.; Zheng, R.; Wang, L.; Yang, H.; et al. MicroRNA-27a controls the intracellular survival of Mycobacterium tuberculosis by regulating calcium-associated autophagy. Nat. Commun. 2018, 9, 4295.
- Ding, S.; Qu, Y.; Yang, S.; Zhao, Y.; Xu, G. Novel miR-1958 promotes Mycobacterium tuberculosis survival in RAW264.7 cells by inhibiting autophagy via Atg5. J. Microbiol. Biotechnol. 2019, 29, 989–998.
- Yuan, Q.; Chen, H.; Yang, Y.; Fu, Y.; Yi, Z. miR-18a promotes Mycobacterial survival in macrophages via inhibiting autophagy by downregulation of ATM. J. Cell. Mol. Med. 2020, 24, 2004–2012.
- Ouimet, M.; Koster, S.; Sakowski, E.; Ramkhelawon, B.; Van Solingen, C.; Oldebeken, S.; Karunakaran, D.; Portal-celhay, C.; Sheedy, F.J.; Ray, T.D.; et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat. Immunol. 2016, 17, 677–686.
- Martinez-Medina, M.; Aldeguer, X.; Lopez-Siles, M.; González-Huix, F.; López-Oliu, C.; Dahbi, G.; Bianco, J.E.; Blanco, J.; Garcia-Gil, L.J.; Darfeuille-Michaud, A. Molecular diversity of Escherichia coli in the human gut: New ecological evidence supporting the role of Adherent–Invasive E. coli (AIEC) in Crohn’s disease. Inflamm. Bowel Dis. 2009, 15, 872–882.
- Larabi, A.; Dalmasso, G.; Delmas, J.; Barnich, N.; Nguyen, H.T.T. Exosomes transfer miRNAs from cell-to-cell to inhibit autophagy during infection with Crohn’s disease-associated Adherent–Invasive E. coli. Gut Microbes 2020, 11, 1677–1694.
- Herkt, C.E.; Caffrey, B.E.; Surmann, K.; Blankenburg, S.; Salazar, G.; Jung, A.L.; Herbel, S.M.; Hoffmann, K.; Schulte, L.N.; Chen, W.; et al. A microRNA network controls Legionella pneumophila replication in human macrophages via LGALS8 and MX1. MBio 2020, 11, e03155-19.
- Chowdhury, S.R.; Reimer, A.; Sharan, M.; Kozjak-Pavlovic, V.; Eulalio, A.; Prusty, B.K.; Fraunholz, M.; Karunakaran, K.; Rudel, T. Chlamydia preserves the mitochondrial network necessary for replication via microRNA-dependent inhibition of fission. J. Cell Biol. 2017, 216, 1071–1089.
- Spier, A.; Stavru, F.; Cossart, P. Interaction between intracellular bacterial pathogens and host cell mitochondria. Microbiol. Spectr. 2019, 7, BAI-0016-2019.
- Sunkavalli, U.; Aguilar, C.; Silva, R.J.; Sharan, M.; Cruz, A.R.; Tawk, C.; Maudet, C.; Mano, M.; Eulalio, A. Analysis of host microRNA function uncovers a role for miR-29b-2-5p in Shigella capture by filopodia. PLoS Pathog. 2017, 13, e1006327.
- Maudet, C.; Mano, M.; Sunkavalli, U.; Sharan, M.; Giacca, M.; Förstner, K.U.; Eulalio, A. Functional high-throughput screening identifies the miR-15 microRNA family as cellular restriction factors for Salmonella infection. Nat. Commun. 2014, 5, 4718.
- Aguilar, C.; Costa, S.; Maudet, C.; Vivek-Ananth, R.P.; Zaldívar-López, S.; Garrido, J.J.; Samal, A.; Mano, M.; Eulalio, A. Reprogramming of microRNA expression via E2F1 downregulation promotes Salmonella infection both in infected and bystander cells. Nat. Commun. 2021, 12, 3392.
- Hu, Z.Q.; Rao, C.L.; Tang, M.L.; Zhang, Y.; Lu, X.X.; Chen, J.G.; Mao, C.; Deng, L.; Li, Q.; Mao, X.H. Rab32 GTpase, as a direct target of miR-30b/c, controls the intracellular survival of Burkholderia pseudomallei by regulating phagosome maturation. PLoS Pathog. 2019, 15, e1007879.
- Fang, Y.; Chen, H.; Hu, Y.; Li, Q.; Hu, Z.; Ma, T.; Mao, X. Burkholderia pseudomallei-derived miR-3473 enhances NF-κB via targeting TRAF3 and is associated with different inflammatory responses compared to Burkholderia thailandensis in murine macrophages. BMC Microbiol. 2016, 16, 283.
- Du, C.T.; Gao, W.; Ma, K.; Yu, S.X.; Li, N.; Yan, S.Q.; Zhou, F.H.; Liu, Z.Z.; Chen, W.; Lei, L.C.; et al. MicroRNA-146a deficiency protects against Listeria monocytogenes infection by modulating the gut microbiota. Int. J. Mol. Sci. 2018, 19, 993.
- Wang, Z.; Brandt, S.; Medeiros, A.; Wang, S.; Wu, H.; Dent, A.; Serezani, C.H. MicroRNA 21 Is a homeostatic regulator of macrophage polarization and prevents prostaglandin e2 -mediated M2 generation. PLoS ONE 2015, 10, e0115855.
- Johnston, D.G.W.; Kearney, J.; Zasłona, Z.; Williams, M.A.; O’Neill, L.A.J.; Corr, S.C. MicroRNA-21 limits uptake of Listeria monocytogenes by macrophages to reduce the intracellular niche and control infection. Front. Cell. Infect. Microbiol. 2017, 7, 201.
- Zhang, J.; Yuan, J.; Wang, L.; Zheng, Z.; Ran, H.; Liu, F.; Li, F.; Tang, X.; Zhang, J.; Ni, Q.; et al. MiR-26a targets EphA2 to resist intracellular Listeria monocytogenes in macrophages. Mol. Immunol. 2020, 128, 69–78.
- Subbarayal, P.; Karunakaran, K.; Winkler, A.C.; Rother, M.; Gonzalez, E.; Meyer, T.F.; Rudel, T. EphrinA2 Receptor (EphA2) is an invasion and intracellular signaling receptor for Chlamydia trachomatis. PLoS Pathog. 2015, 11, e1004846.
- Bravo-santano, N.; Stölting, H.; Cooper, F.; Bileckaja, N.; Majstorovic, A.; Ihle, N.; Mateos, L.M.; Calle, Y.; Behrends, V.; Letek, M. Host-directed kinase inhibitors act as novel therapies against intracellular Staphylococcus aureus. Sci. Rep. 2019, 9, 4876.
- Iannaccone, M.; Dorhoi, A.; Kaufmann, S.H.E. Host-directed therapy of tuberculosis: What is in it for microRNA? Expert Opin. Ther. Targets 2014, 18, 491–494.
- Man, D.K.W.; Chow, M.Y.T.; Casettari, L.; Gonzalez-Juarrero, M.; Lam, J.K.W. Potential and development of inhaled RNAi therapeutics for the treatment of pulmonary tuberculosis. Adv. Drug Deliv. Rev. 2016, 102, 21–32.
- Deng, Y.; Wang, C.C.; Choy, K.W.; Du, Q.; Chen, J.; Wang, Q.; Li, L.; Chung, T.K.H.; Tang, T. Therapeutic potentials of gene silencing by RNA interference: Principles, challenges, and new strategies. Gene 2014, 538, 217–227.
- Pecot, C.V.; Calin, G.A.; Coleman, R.L.; Lopez-Berestein, G.; Sood, A.K. RNA interference in the clinic: Challenges and future directions. Nat. Rev. Cancer 2011, 11, 59–67.
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther.-Nucleic Acids 2015, 4, e252.
- Rosas-taraco, A.G.; Higgins, D.M.; Sánchez-campillo, J.; Lee, E.J.; Orme, I.M.; González-juarrero, M. Local pulmonary immunotherapy with siRNA targeting TGFβ1 enhances antimicrobial capacity in Mycobacterium tuberculosis infected mice. Tuberculosis 2011, 91, 98–106.
- Drury, R.E.; O’Connor, D.; Pollard, A.J. The clinical application of MicroRNAs in infectious disease. Front. Immunol. 2017, 8, 1182.
- Jamalkhah, M.; Asaadi, Y.; Azangou-Khyavy, M.; Khanali, J.; Soleimani, M.; Kiani, J.; Arefian, E. MSC-derived exosomes carrying a cocktail of exogenous interfering RNAs an unprecedented therapy in era of COVID-19 outbreak. J. Transl. Med. 2021, 19, 164.
- Gebert, L.F.R.; Rebhan, M.A.E.; Crivelli, S.E.M.; Denzler, R.; Stoffel, M.; Hall, J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014, 42, 609–621.
- Janssen, H.L.A.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694.
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24.
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008.
- Cullis, P.R.; Hope, M.J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 2017, 25, 1467–1475.
- Guo, P.; Coban, O.; Snead, N.M.; Trebley, J.; Hoeprich, S.; Guo, S.; Shu, Y. Engineering RNA for targeted siRNA delivery and medical application. Adv. Drug Deliv. Rev. 2010, 62, 650–666.
- Akuma, P.; Okagu, O.D.; Udenigwe, C.C. Naturally Occurring Exosome Vesicles as Potential Delivery Vehicle for Bioactive Compounds. Front. Sustain. Food Syst. 2019, 3, 23.
- Mathiyalagan, P.; Sahoo, S. Exosomes-based gene therapy for MicroRNA delivery. Methods Mol. Biol. 2017, 1521, 139–152.
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228.
- Qiao, L.; Hu, S.; Huang, K.; Su, T.; Li, Z.; Vandergriff, A.; Cores, J. Theranostics tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs. Theranostics 2020, 10, 3474–3487.
- Alvarez-erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345.
- Wang, J.; Dong, Y.; Li, Y.; Li, W.; Cheng, K.; Qian, Y.; Xu, G. Designer exosomes for active targeted chemo-photothermal synergistic tumor therapy. Adv. Func. Mat. 2018, 28, 1707360.
- Matsuki, Y.; Yanagawa, T.; Sumiyoshi, H.; Yasuda, J. Modification of exosomes with carbonate apatite and a glycan polymer improves transduction ef fi ciency and target cell selectivity. Biochem. Biophys. Res. Commun. 2021, 583, 93–99.
- Schorey, J.S.; Harding, C.V.; Schorey, J.S.; Harding, C. V Extracellular vesicles and infectious diseases : New complexity Extracellular vesicles and infectious diseases : New complexity to an old story. J. Clin. Investig. 2016, 126, 1181–1189.
- Hosseini, H.M.; Ali, A.; Fooladi, I.; Nourani, M.R.; Ghanezadeh, F. The role of exosomes in infectious diseases. Inflamm. Allergy-Drug Targets 2013, 12, 29–37.
- Jones, L.B.; Bell, C.R.; Bibb, K.E.; Gu, L.; Coats, M.T.; Matthews, Q.L. Pathogens and their effect on exosome biogenesis and composition. Biomedicines 2018, 6, 79.
- Neupane, K.R.; Mccorkle, J.R.; Kopper, T.J.; Lakes, J.E.; Aryal, S.P.; Abdullah, M.; Snell, A.A.; Gensel, J.C.; Kolesar, J.; Richards, C.I. Macrophage-engineered vesicles for therapeutic delivery and bidirectional reprogramming of immune cell polarization. ACS Omega 2021, 6, 3847–3857.
- Alipoor, S.D.; Mortaz, E.; Tabarsi, P.; Farnia, P.; Mirsaeidi, M.; Garssen, J.; Movassaghi, M.; Adcock, I.M. Bovis Bacillus Calmette-Guerin (BCG) infection induces exosomal miRNA release by human macrophages. J. Transl. Med. 2017, 15, 105.
- Mosquera-Heredia, M.I.; Morales, L.C.; Vidal, O.M.; Barceló, E.; Silvera-Redondo, C.; Vélez, J.I.; Garavito-Galofre, P. Exosomes: Potential disease biomarkers and new therapeutic targets. Biomedicines 2021, 9, 1061.
- Barile, L.; Vassalli, G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 2017, 174, 63–78.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
672
Revisions:
2 times
(View History)
Update Date:
25 Mar 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




