Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | YUANZHE LI | + 1423 word(s) | 1423 | 2022-02-22 06:45:27 | | | |
| 2 | YUANZHE LI | Meta information modification | 1423 | 2022-03-23 09:27:56 | | | | |
| 3 | Catherine Yang | Meta information modification | 1423 | 2022-03-23 09:30:13 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Li, Y. Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Encyclopedia. Available online: https://encyclopedia.pub/entry/20908 (accessed on 07 February 2026).
Li Y. Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Encyclopedia. Available at: https://encyclopedia.pub/entry/20908. Accessed February 07, 2026.
Li, Yuanzhe. "Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry" Encyclopedia, https://encyclopedia.pub/entry/20908 (accessed February 07, 2026).
Li, Y. (2022, March 23). Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. In Encyclopedia. https://encyclopedia.pub/entry/20908
Li, Yuanzhe. "Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry." Encyclopedia. Web. 23 March, 2022.
Copy Citation
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) is an advanced technique that uses minimum fragmented ions from complex molecules for mass spectrometry (MS) analysis (tissue profiling by mass spectrometry). It is able to analyze spatially resolved tissue or tumor sections at the molecular level. It has become a valuable tool for tumor and tissue imaging, due to its ease of operation and high mass resolution, but it still has vast room for development in the instrumentation of larger proteins in some tissues.
matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS)
1. Introduction
Matrix-assisted laser desorption/ionization (MALDI) is an ionization process that utilizes a laser irradiation absorbing matrix to produce minimum fragmented ions from complex molecules for mass spectrometry (MS) analysis (tissue profiling by mass spectrometry). In 1994, the first use of MALDI-MS in an imaging context, for matrix crystals containing the neuropeptide P, was reported by Spengler et al. [1] (Small molecule MALDI MS imaging: Current technologies and future challenges). With the development of MALDI-MS, its application areas have rapidly expanded. MALDI-MS is a powerful analytical technique that combines the high sensitivity and selectivity of mass spectrometry with the spatial distribution data of molecules in tumors or tissues [2]. It is target free; therefore, it does not need to choose a certain protein analyte ahead of time. Although biased detections may occur sometimes, it is fully multiplex, and it measures all ions from a tissue sample simultaneously. Therefore, using MALDI-MS, protein localization can be determined, with molecular specificity, directly from tissue parts [1]. Furthermore, since the MALDI laser does not ablate any of the tissue, and molecules are desorbed from the surface, cellular and molecular integrity is maintained, with minimal effects on most of the cells. Oppenheimer’s report provides an overview of the workflow of the analysis, providing a detailed introduction of the instrumentation and applications to clinical oncology and pharmaceutical development [3]. As a general outline, the analysis uses a matrix that absorbs energy at the irradiating laser’s wavelength. The sample molecule is combined with the matrix, then dried on an electrically conductive glass slide. Matrix–analyte co-crystals develop during the drying process. Such crystals are then exposed to UV laser light, allowing the sample molecule to desorb and ionize. Then, in a time-of-flight (TOF) mass analyzer, a mass-to-charge (m/z) ratio is generated [4]. Finally, a 2D ion intensity map can be constructed based on the signals obtained at x, y coordinates. It should be noted that, due to the limited coverage of various aspects, such as data analysis techniques and other types of detectors, the TOF mass analyzer within this paper only represents one representative example of MALDI imaging.
2. Principle of MALDI-MS for Tissue or Tumor Imaging
The general workflow of how MALDI-MS works to produce images of tumors and tissue is shown in Figure 1 [5]. Firstly, a sliced tissue or tumor is pretreated, and a matrix is needed to cover it. Then, the mass spectrometer analyzes the tissue specimen (with a spatial resolution varying from approximately 200 μm to 20 μm), producing a mass spectrum for each measurement spot.
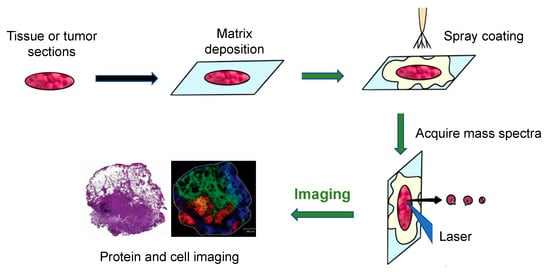
Figure 1. The principal workflow of the MALDI imaging and profiling experiments [5].
An Ultraflex II MALDI-TOF-MS (Bruker Daltonics, Billerica, MA, USA), equipped with a SmartBeam TM laser, can be used for tissue or tumor imaging. Using this machine, a total of 400 laser shots are obtained for each matrix spot, in increments of 50 shots, starting at the middle of each spot and randomly rastering at various locations inside the spot. An evaluation of the relative coordinates of each matrix spot from the optical picture of the MALDI plate is required for automated acquisition of the profile. Then, the x, y coordinates of each spot are written on a file in the native target geometry of the mass spectrometer control program. Three positions from the outer corners of the array are chosen for final alignment of the plate [3]. Finally, plotting the intensities of each signal at its x, y coordinates can create a two-dimensional morphological image of the ion profiles. Since the sliced tissue or tumor can be stained, the observed mass signals can then be observed as color intensity maps and can be used to investigate molecules in certain cell types. Using these colored signals, the distribution of different molecules in the tissues can be visualized [6].
3. Limitations and Future Applications
MALDI-MS analyses are successful for most tumors and tissues. Due to the limitation of the article length, the protocol described above is only suitable for peptides (proteins), but is not suitable for small molecules. Similarly, the method that describes rinsing with ethanol and adding trypsin are optional for proteins targeted by MALDI-MS, but not for small molecules. Moreover, it is less effective at detecting larger proteins in some tissues because the mass range of such a methodology is limited by laser ionization and ablation processes, which lead to the fragmentation of larger molecules, such as cytokines, growth factors, enzymes and receptors that have molecular weights exceeding 25 kDa, and because MALDI-MS is vulnerable to detector saturation when studying complicated mixtures. This limitation could be overcome by using a high mass detector, but this would generate a significant chemical background, which would affect any losses of sensitivity. Hence, a high mass detector that can withstand the high chemical background of MALDI-MS is required [7]. In addition, due to the limitations of this mini-review, only limited coverage of various aspects, such as data analysis techniques and detector coverage, is included within this paper. Such examples may only indicate the distribution of three molecules, rather than illustrating how to use the technique to diagnose via tumor subtyping.
3.1. Diagnostic and Prognostic Assessment in Clinical Pathology
Recently, MALDI-MS has been applied to cancer research, including human non-small-cell lung tumors, gliomas, breast cancer and ovarian tumors. The general method employed by these studies is to compare the mass spectral characteristics (m/z peaks) with a range of patient data, to classify specific molecular alterations related to disease progression [8]. The spatial proteomic characterization of tissue and tumor recognition contributes to better diagnoses and individual predictive trends of therapy response [9].
Generally, 1 μL of extracted serum peptide may be mixed with 1 μL of saturated α-cyano-4-hydroxycinnamic acid matrix (dissolved with 0.1% trifluoroacetic acid, 50% acetonitrile), spotted on a target plate and dried at room temperature; the target plate is placed in the mass spectrometer; the instrument is calibrated with standards, and then the standards are detected to obtain a mass-to-charge ratio (m/z) peptide peak, consisting of different mass-to-charge ratios (m/z). The mass spectra of the peptide peaks with different mass-to-charge ratios (m/z) are then obtained. In order to avoid system errors and human errors, standards (peptide mixtures) are used before each specimen is tested, and the specimens are only tested when the results are consistent with the composition of the standards, indicating that the test system is working properly, thus ensuring reliable and reproducible results.
One segment example of lung cancer tissue, displaying various areas identified by the distinct molecular content of the tissue, is shown in the bottom right of Figure 3 [5]. The left side of the figure shows the H&E staining of the segment after MALDI-MS analysis. The right side of the figure shows a fibrotic region of the tumor cells (m/z = 1117.1, yellow), a tumor region (m/z = 1822.5, red), a non-tumor region (m/z = 1530.4, blue), and a peritumoral inflammatory field (m/z = 1429.1, green). This example illustrates how to use the technique to diagnose disease.
3.2. Tumor Removal
In clinical oncology, completely removing tumors is crucial. Research shows that some of the tumor’s molecular features are represented in histologically healthy tissue neighboring the tumors, due to molecular modifications before phenotypic changes. MALDI-MS can aid in the interpretation of alterations in tumors and neighboring healthy tissues’ environments, as well as provide an approximation of how far these changes extend beyond the histologically defined margins [10]. As a result, the tumors can be resected completely.
3.3. Drug Development
MALDI-MS can be used to investigate the distribution of drugs in human and animal tumor tissues [11]. This technique has improved selectivity and sensitivity, which is ideal for analyzing how effective drugs are and for improving drug design. One study used MALDI-MS to deliver paclitaxel to a mouse with a tumor. Paclitaxel (PTX) was combined with micelle (NK105) and delivered to the tumor tissue of the mouse. Using MALDI-MS, this section was compared with the tumor section treated with PTX alone, as well as the untreated mouse. The PTX concentration delivered from NK105 was significantly higher than the free PTX and untreated tumor tissue, meaning that the anticancer efficacy of NK105 is higher than PTX alone [12].
References
- Gessel, M.; Norris, J.; Caprioli, R.M. MALDI imaging mass spectrometry: Spatial molecular analysis to enable a new age of discovery. J. Proteom. 2014, 107, 71–82.
- Hillenkamp, F.; Karas, M.; Beavis, R.C.; Chait, B.T. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal. Chem. 1991, 63, 1193–1203.
- Oppenheimer, S.R.; Mi, D.; Sanders, M.E.; Caprioli, R.M. Molecular Analysis of Tumor Margins by MALDI Mass Spectrometry in Renal Carcinoma. J. Proteome Res. 2010, 9, 2182–2190.
- Du, Y.; Du, Y.; Cui, M.; Liu, Z. Characterization of the Noncovalent Interactions between Lysozyme and Panaxadiol Glycosides by Intensity-Fading—Matrix-Assisted Laser Desorption Ionization—Mass Spectrometry (IF-MALDI-MS). Anal. Lett. 2021, 54, 2387–2394.
- Balluff, B.; Schöne, C.; Höfler, H.; Walch, A. MALDI imaging mass spectrometry for direct tissue analysis: Technological advancements and recent applications. Histochem. Cell Biol. 2011, 136, 227–244.
- Norris, J.; Caprioli, R.M. Analysis of Tissue Specimens by Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry in Biological and Clinical Research. Chem. Rev. 2013, 113, 2309–2342.
- Yang, J.; Caprioli, R.M. Matrix pre-coated targets for high throughput MALDI imaging of proteins. Biol. Mass Spectrom. 2014, 49, 417–422.
- Thurner, G.C.; Debbage, P. Molecular imaging with nanoparticles: The dwarf actors revisited 10 years later. Histochem. Cell Biol. 2018, 150, 733–794.
- Schaepe, K.; Bhandari, D.R.; Werner, J.; Henss, A.; Pirkl, A.; Kleine-Boymann, M.; Rohnke, M.; Wenisch, S.; Neumann, E.; Janek, J.; et al. Imaging of Lipids in Native Human Bone Sections Using TOF–Secondary Ion Mass Spectrometry, Atmospheric Pressure Scanning Microprobe Matrix-Assisted Laser Desorption/Ionization Orbitrap Mass Spectrometry, and Orbitrap–Secondary Ion Mass Spectrometry. Anal. Chem. 2018, 90, 8856–8864.
- Wu, N.; Jiao, L.; Bütikofer, M.; Zeng, Z.; Zenobi, R. High-Mass Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry for Absolute Quantitation of Noncovalent Protein–Protein Binding Interactions. Anal. Chem. 2021, 93, 10982–10989.
- Römpp, A.; Spengler, B. Mass spectrometry imaging with high resolution in mass and space. Histochem. Cell Biol. 2013, 139, 759–783.
- Yasunaga, M.; Furuta, M.; Ogata, K.; Koga, Y.; Yamamoto, Y.; Takigahira, M.; Matsumura, Y. The significance of microscopic mass spectrometry with high resolution in the visualisation of drug distribution. Sci. Rep. 2013, 3, 3050.
More
Information
Subjects:
Microbiology; Instruments & Instrumentation
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Entry Collection:
Biopharmaceuticals Technology
Revisions:
3 times
(View History)
Update Date:
23 Mar 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




