
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Pablo Yagupsky | + 2335 word(s) | 2335 | 2022-03-21 04:36:50 | | | |
| 2 | Peter Tang | Meta information modification | 2335 | 2022-03-22 02:12:12 | | |
Video Upload Options
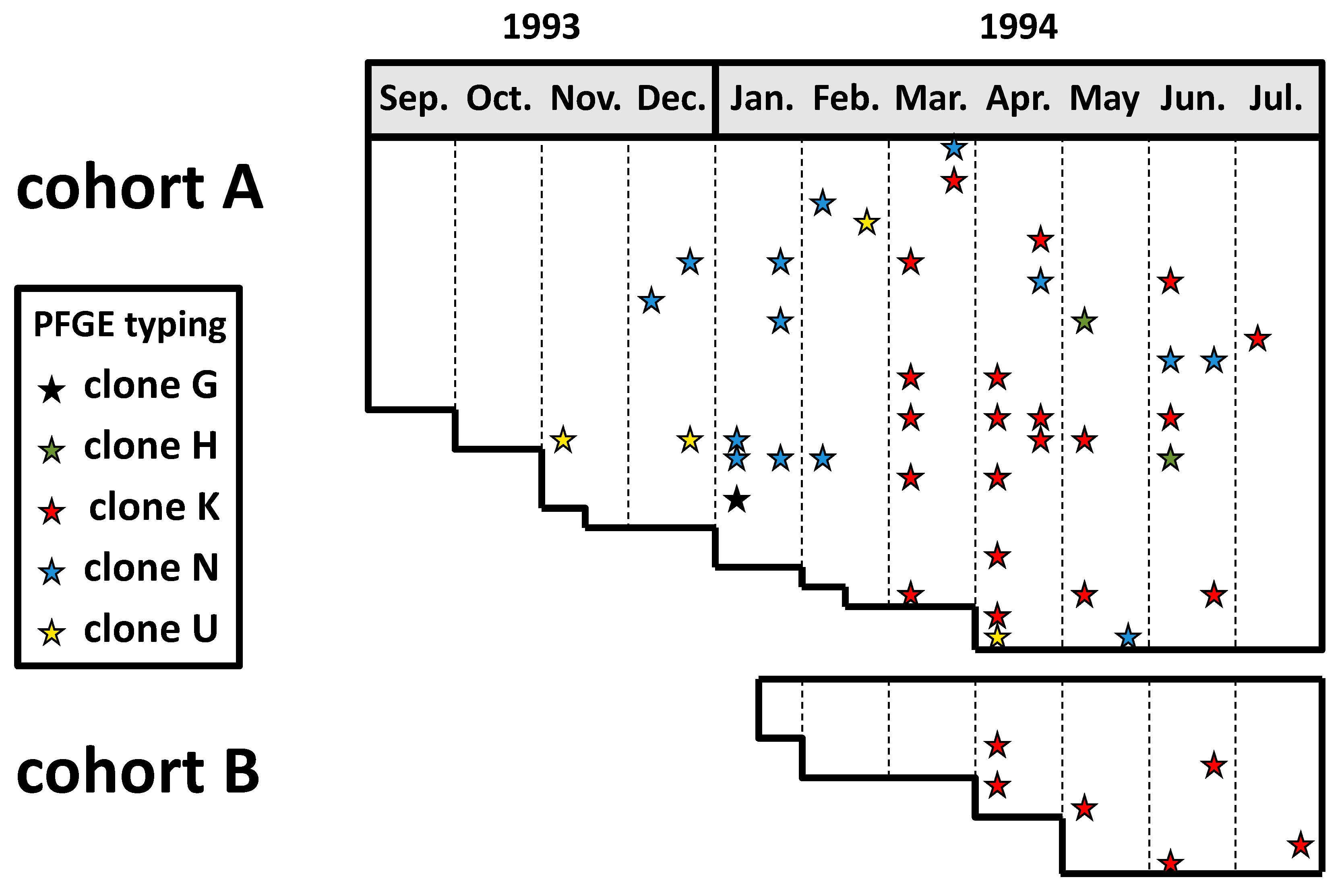
Kingella kingae colonizes the oropharynx after the second life semester, and its prevalence reaches 10% between the ages of 12 and 24 months, declining thereafter as children reach immunological maturity. Kingella kingae colonization is characterized by the periodic substitution of carried organisms by new strains. Whereas some strains frequently colonize asymptomatic children but are rarely isolated from diseased individuals, others are responsible for most invasive infections worldwide, indicating enhanced virulence. The colonized oropharyngeal mucosa is the source of child-to-child transmission, and daycare attendance is associated with a high carriage rate and increased risk of invasive disease. Kingella kingae elaborates a potent repeat-in-toxin (RTXA) that lyses epithelial, phagocytic, and synovial cells. This toxin breaches the epithelial barrier, facilitating bloodstream invasion and survival and the colonization of deep body tissues.
1. Introduction
2. Kingella kingae: An Oropharyngeal Resident
3. Mechanism of Colonization
4. Immunity to Colonization and Infection
5. Colonization and Transmission
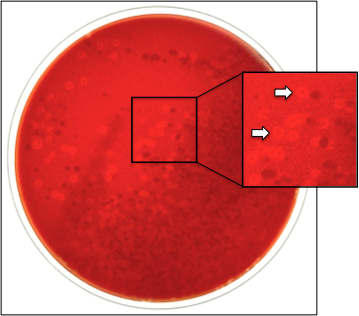
6. Detection of K. kingae Colonization
References
- García-Rodríguez, J.Á.; Martínez, M.J.F. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J. Antimicrob. Chemother. 2002, 50 (Suppl. S2), 59–74.
- Weintraub, A. Immunology of bacterial polysaccharide antigens. Carbohydr. Res. 2003, 338, 2539–2547.
- Samuelson, A.; Freijd, A.; Jonasson, J.; Lindberg, A.A. Turnover of nonencapsulated Haemophilus influenzae in the nasopharynges of otitis-prone children. J. Clin. Microbiol. 1995, 33, 2027–2031.
- Sethi, S.; Hill, S.L.; Murphy, T.F. Serum antibodies to outer membrane proteins (OMPs) of Moraxella (Branhamella) catarrhalis in patients with bronchiectasis: Identification of OMP B1 as an important antigen. Infect. Immun. 1995, 63, 1516–1520.
- Yagupsky, P.; Dagan, R.; Prajgrod, F.; Merires, M. Respiratory carriage of Kingella kingae among healthy children. Pediatr. Infect. Dis. J. 1995, 14, 673–677.
- Amit, U.; Flaishmakher, S.; Dagan, R.; Porat, N.; Yagupsky, P. Age-dependent carriage of Kingella kingae in young children and turnover of colonizing strains. J. Pediatr. Infect. Dis. Soc. 2013, 3, 160–162.
- Kehl-Fie, T.E.; Miller, S.E.; St. Geme, J.W., 3rd. Kingella kingae Expresses type IV Pili that mediate adherence to respiratory epithelial and synovial cells. J. Bacteriol. 2008, 190, 7157–7163.
- Porsch, E.A.; Johnson, M.D.L.; Broadnax, A.D.; Garrett, C.K.; Redinbo, M.R.; St. Geme, J.W., 3rd. Calcium Binding Properties of the Kingella kingae PilC1 and PilC2 proteins have differential effects on type IV pilus-mediated adherence and twitching motility. J. Bacteriol. 2012, 195, 886–895.
- Kehl-Fie, T.E.; Porsch, E.A.; Miller, S.E.; St. Geme, J.W., 3rd. Expression of Kingella kingae type IV pili is regulated by θ54, PilS, and PilR. J. Bacteriol. 2009, 191, 4976–4986.
- Kehl-Fie, T.E.; Porsch, E.A.; Yagupsky, P.; Grass, E.A.; Obert, C.; Benjamin, D.K., Jr.; St. Geme, J.W., 3rd. Examination of type IV pilus expression and pilus-associated phenotypes in Kingella kingae clinical isolates. Infect. Immun. 2010, 78, 1692–1699.
- Porsch, E.A.; Kehl-Fie, T.E.; St. Geme, J.W., 3rd. Modulation of Kingella kingae Adherence to human epithelial cells by type IV Pili, capsule, and a novel trimeric autotransporter. mBio 2012, 3, e00372-12.
- Yagupsky, P. Kingella kingae: Carriage, transmission, and disease. Clin. Microbiol. Rev. 2015, 28, 54–79.
- Slonim, A.; Steiner, M.; Yagupsky, P. Immune response to invasive Kingella kingae infections, age-related incidence of disease, and levels of antibody to outer-membrane proteins. Clin. Infect. Dis. 2003, 37, 521–527.
- Starr, K.F.; Porsch, E.A.; Heiss, C.; Black, I.; Azadi, P.; St. Geme, J.W., 3rd. Characterization of the Kingella kingae polysaccharide capsule and exopolysaccharide. PLoS ONE 2013, 8, e75409.
- Muñoz, V.L.; Porsch, E.A.; St. Geme, J.W., 3rd. Kingella kingae surface polysaccharides promote resistance to neutrophil phagocytosis and killing. mBio 2019, 10, e00631-19.
- Muñoz, V.L.; Porsch, E.A.; St. Geme, J.W., 3rd. Kingella kingae surface polysaccharides promote resistance to human serum and virulence in a juvenile rat model. Infect. Immun. 2018, 86, e00100-18.
- Slonim, A.; Walker, E.S.; Mishori, E.; Porat, N.; Dagan, R.; Yagupsky, P. Person-to-person transmission of Kingella kingae among day care center attendees. J. Infect Dis. 1998, 78, 1843–1846.
- Amit, U.; Dagan, R.; Porat, N.; Trefler, R.; Yagupsky, P. Epidemiology of invasive Kingella kingae infections in two distinct pediatric populations cohabiting in one geographic area. Pediatr. Infect. Dis. J. 2012, 31, 415–417.
- Yagupsky, P.; Weiss-Salz, I.; Fluss, R.; Freedman, L.; Peled, N.; Trefler, R.; Porat, N.; Dagan, R. Dissemination of Kingella kingae in the community and long-term persistence of invasive clones. Pediatr. Infect. Dis. J. 2009, 28, 707–710.
- Yagupsky, P.; Merires, M.; Bahar, J.; Dagan, R. Evaluation of a novel vancomycin-containing medium for primary isolation of Kingella kingae from upper respiratory tract specimens. J. Clin. Microbiol. 1995, 33, 426–427.
- Basmaci, R.; Ilharreborde, B.; Bidet, P.; Doit, C.; Lorrot, M.; Mazda, K.; Bingen, E.; Bonacorsi, S. Isolation of Kingella kingae in the oropharynx during K. kingae arthritis on children. Clin. Microbiol. Infect. 2012, 18, e134–e136.
- Yagupsky, P.; El Houmami, N.; Fournier, P.-E. Outbreaks of invasive Kingella kingae infections in daycare facilities: Approach to investigation and management. J. Pediatr. 2017, 182, 14–20.
- Seña, A.C.; Seed, P.; Nicholson, B.; Joyce, M.; Cunningham, C.K. Kingella kingae endocarditis and a cluster investigation among daycare attendeES. Pediatr. Infect. Dis. J. 2010, 29, 86–88.
- Chometon, S.; Benito, Y.; Chaker, M.; Boisset, S.S.; Ploton, C.; Bérard, J.; Vandenesch, F.; Freydiere, A.M. Specific real-time polymerase chain reaction places Kingella kingae as the most common cause of osteoarticular infections in young children. Pediatr. Infect. Dis. J. 2007, 26, 377–381.
- Ceroni, D.; Anderson della Llana, R.; Kherad, O.; Lascombes, P.; Renzi, G.; Manzano, S.; Cherkaoui, A.; Schrenzel, J. Comparing the oropharyngeal colonizationisation density of Kingella kingae between asymptomatic carriers and children with invasive osteoarticular infections. Pediatr. Infect. Dis. J. 2013, 32, 412–414.
- Rosey, A.L.; Abachin, E.; Quesnes, G.; Cadilhac, C.; Pejin, Z.; Glorion, C.; Berche, P.; Ferroni, A. Development of a braod-range 16S rDNA real-time PCR for the diagnosis of septic arthritis in children. J. Microbiol. Methods. 2007, 68, 88–93.
- Moumile, K.; Merckx, J.; Glorion, C.; Berche, P.; Ferroni, A. Osteoarticular infections caused by Kingella kingae in children: Contribution of polymerase chain reaction to the microbiologic diagnosis. Pediatr. Infect. Dis. J. 2003, 22, 837–839.
- El Houmami, N.; Durand, G.A.; Bzdrenga, J.; Darmon, A.; Minodier, P.; Seligmann, H.; Raoult, D.; Fournier, P.-E. A new highly sensitive and specific real-time pcr assay targeting the malate dehydrogenase gene of Kingella kingae and application to 201 pediatric clinical specimens. J. Clin. Microbiol. 2018, 56, e00505-18.
- Ceroni, D.; Cherkaoui, A.; Kaelin, A.; Schrenzel, J. Kingella kingae spondylodiscitis in young children: Toward a new approach for bacteriological investigations? A preliminary report. J. Child. Orthop. 2010, 4, 173–175.
- Juchler, C.; Spyropoulou, V.; Wagner, N.; Merlini, L.; Dhouib, A.; Manzano, S.; Tabard-Fougère, A.; Samara, E.; Ceroni, D. The contemporary bacteriologic epidemiology of osteoarticular infections in children in Switzerland. J. Pediatr. 2018, 194, 190–196.e1.
- Masud, S.; Greenman, J.; Mulpuri, K.; Hasan, M.R.; David, M.; Goldfarb, D.M.; Tilley, P.; Gadkar, V.J.; Al-Rawahi, G.N. Asymptomatic pharyngeal carriage of Kingella kingae among young children in Vancouver, British Columbia, Canada. Pediatr. Infect. Dis. J. 2019, 38, 990–993.
- Lehours, P.; Freydière, A.M.; Richer, O.; Burucoa, C.; Boisset, S.; Lanotte, P.; Prère, M.F.; Ferroni, A.; Lafuente, C.; Vandenesch, F.; et al. The rtxA toxin gene of Kingella kingae: A pertinent target for molecular diagnosis of osteo- articular infections. J. Clin. Microbiol. 2011, 49, 1245–1250.




