
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Natalia Mokshina | + 2653 word(s) | 2653 | 2022-03-17 07:37:12 | | | |
| 2 | Catherine Yang | + 1 word(s) | 2654 | 2022-03-21 09:12:37 | | |
Video Upload Options
Plant proteins with lectin domains play an essential role in plant immunity modulation, but among a plurality of lectins recruited by plants, only a few members have been functionally characterized. The lectin gene expression in flax root tips infected with Fusarium oxysporum was analyzed. For the analysis of flax lectin gene expression, FIBexDB was used, which includes an efficient algorithm for flax gene expression analysis combining gene clustering and coexpression network analysis. Two pools of lectin genes have been revealed: downregulated and upregulated during the infection.
1. Introduction
2. Expression of Flax Genes for Lectins during Fungal Infection in Different Cultivars
In flax root tips, 304 genes with lectin domains were expressed (threshold of TGR (total gene read) value ≥ 16 at least in one sample). According to the cluster analysis performed using FIBexDB, this set of lectin genes was subdivided into three clusters (Figure 1): 63 genes accounted for the cluster where genes in general showed down-regulation of expression in the root samples of all flax genotypes treated with F. oxysporum compared to controls (cluster 1). The expression of 216 genes was upregulated in all flax genotypes treated with F. oxysporum as compared to controls: 146 genes were upregulated in all infected plants independently of the flax genotype (cluster 2). Cluster 3 included 70 genes whose expression in the susceptible cultivars was less pronounced compared to the resistant cultivars and hybrids.
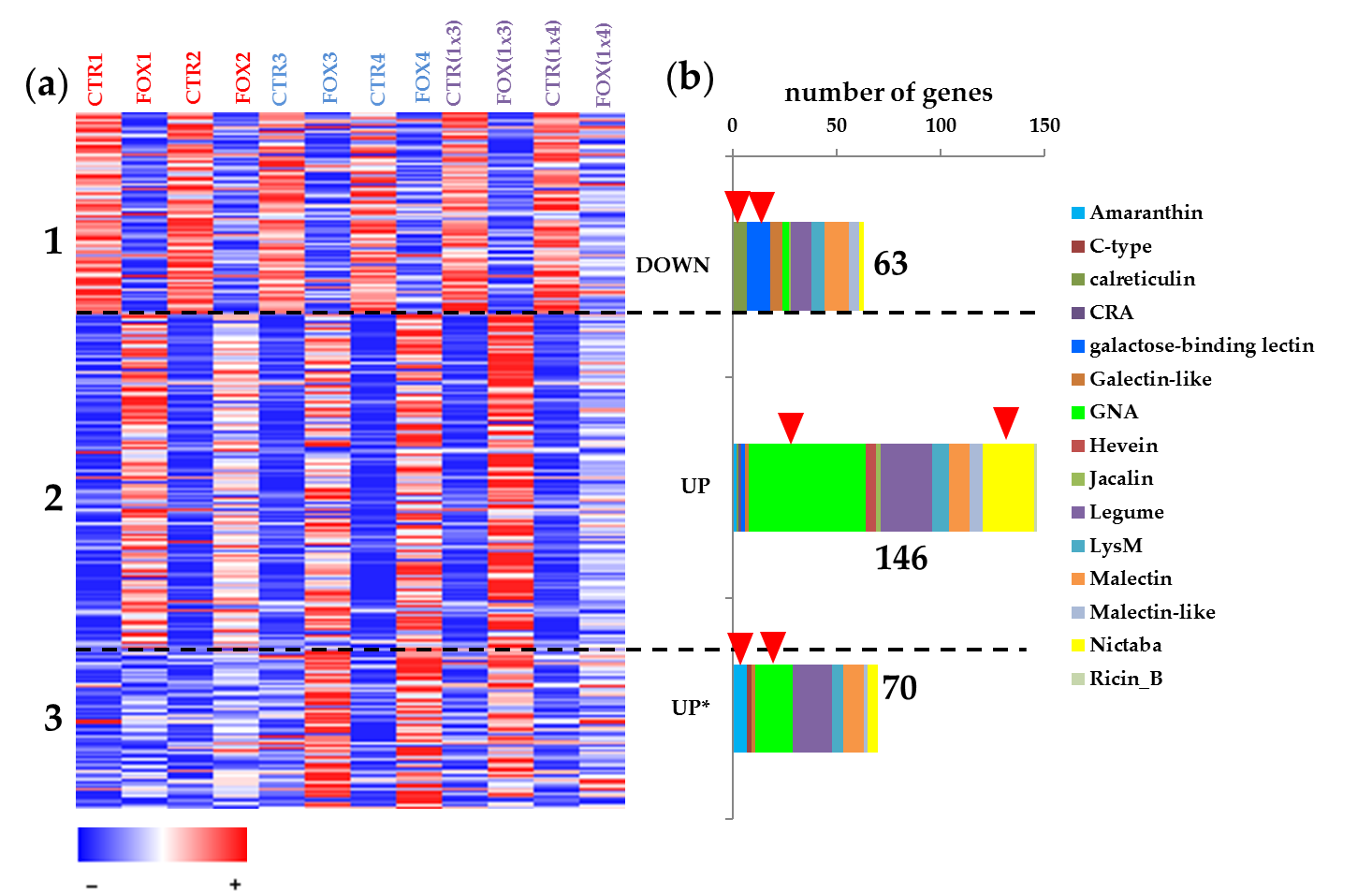
Figure 1. (a) Cluster analysis of 304 genes expressed in flax root tips of different genotypes treated with Fusarium oxysporum. A heatmap was built in FiBexDB. CTR1,2,3,4,(1x3),(1x4)—untreated root tips of different flax genotypes, FOX1,2,3,4,(1x3),(1x4)—root tips of different flax genotypes treated with F. oxysporum. *—genes for lectins upregulated in cultivars resistant to F. oxysporum. (b) Members of different lectin gene families in the analyzed clusters. Red arrows mark family members which were abundant in the analyzed clusters.
3. Genes Encoding Lectins Potentially Involved in the Biosynthesis of Cell Compounds
4. The Avant-Garde of Defense Flax Lectins in Response to Fusarium oxysporum
5. Lectins That Might Determine the Resistance of Flax to F. oxysporum

References
- Jaber, K.; Haubruge, É.; Francis, F. Development of entomotoxic molecules as control agents: Illustration of some protein potential uses and limits of lectins. Biotechnol. Agron. Soc. Environ. 2010, 14, 225–241.
- Newman, M.A.; Sundelin, T.; Nielsen, J.T.; Erbs, G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 2013, 4, 139.
- Singh, P.; Zimmerli, L. Lectin receptor kinases in plant innate immunity. Front. Plant Sci. 2013, 4, 124.
- Santos, A.F.S.; Da Silva, M.D.C.; Napoleão, T.H.; Paiva, P.M.G.; Correia, M.T.S.; Coelho, L.C.B.B. Lectins: Function, structure, biological properties andpotential applications. Curr. Top. Pept. Protein Res. 2014, 15, 41–62.
- Tsaneva, M.; Van Damme, E.J.M. 130 years of Plant Lectin Research. Glycoconj. J. 2020, 37, 533–551.
- Petrova, N.; Nazipova, A.; Gorshkov, O.; Mokshina, N.; Patova, O.; Gorshkova, T. Gene expression patterns for proteins with lectin domains in flax stem tissues are related to deposition of distinct cell wall types. Front. Plant Sci. 2021, 12, 634594.
- Jiang, S.-Y.; Ma, Z.; Ramachandran, S. Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol. Biol. 2010, 10, 79.
- Lannoo, N.; Van Damme, E.J.M. Lectin domains at the frontiers of plant defense. Front. Plant Sci. 2014, 5, 397.
- Alexey A. Dmitriev; George S. Krasnov; Tatiana A. Rozhmina; Roman O. Novakovskiy; Anastasiya V. Snezhkina; Maria S. Fedorova; Olga Yu. Yurkevich; Olga V. Muravenko; Nadezhda L. Bolsheva; Anna V. Kudryavtseva; et al.Nataliya V. Melnikova Differential gene expression in response to Fusarium oxysporum infection in resistant and susceptible genotypes of flax (Linum usitatissimum L.). BMC Plant Biology 2017, 17, 29-40, 10.1186/s12870-017-1192-2.
- Natalia Mokshina; Oleg Gorshkov; Hironori Takasaki; Hitomi Onodera; Shingo Sakamoto; Tatyana Gorshkova; Nobutaka Mitsuda; FIBexDB: a new online transcriptome platform to analyze development of plant cellulosic fibers. New Phytologist 2021, 231, 512-515, 10.1111/nph.17405.
- Leach, M.R.; Williams, D.B. Calnexin and Calreticulin, Molecular Chaperones of the Endoplasmic Reticulum. In Calreticulin. Molecular Biology Intelligence; Eggleton, P., Michalak, M., Eds.; Springer: Boston, MA, USA, 2003; pp. 49–62.
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485.
- Pröbsting, M.; Schenke, D.; Hossain, R.; Häder, C.; Thurau, T.; Wighardt, L.; Schuster, A.; Zhou, Z.; Ye, W.; Rietz, S.; et al. Loss of function of CRT1a (calreticulin) reduces plant susceptibility to Verticillium longisporum in both Arabidopsis thaliana and oilseed rape (Brassica napus). Plant Biotech. J. 2020, 18, 2328–2344.
- Nouri, M.Z.; Hiraga, S.; Komatsu, S. Characterization of calnexin in soybean roots and hypocotyls under osmotic stress. Phytochemistry 2012, 74, 20–29.
- Heilmann, I.; Shin, J.; Huang, J.; Perera, I.Y.; Davies, E. Transient dissociation of polyribosomes and concurrent recruitment of calreticulin and calmodulin transcripts in gravistimulated maize pulvini. Plant Physiol. 2001, 127, 1193–1203.
- Sampedro, J.; Gianzo, C.; Iglesias, N.; Guitián, E.; Revilla, G.; Zarra, I. AtBGAL10 Is the Main Xyloglucan β-Galactosidase in Arabidopsis, and Its Absence Results in Unusual Xyloglucan Subunits and Growth Defects. Plant Physiol. 2012, 158, 1146–1157.
- McCarthy, A.A.; McCarthy, J.G. The structure of two N-methyltransferases from the caffeine biosynthetic pathway. Plant Physiol. 2007, 144, 879–889.
- Miao, Y.; Li, H.Y.; Shen, J.; Wang, J.; Jiang, L. QUASIMODO 3 (QUA3) is a putative homogalacturonan methyltransferase regulating cell wall biosynthesis in Arabidopsis suspension-cultured cells. J. Exp. Bot. 2011, 62, 5063–5078.
- Willats, W.G.; Orfila, C.; Limberg, G.; Buchholt, H.C.; van Alebeek, G.J.; Voragen, A.G.; Marcus, S.; Christensen, T.; Mikkelsen, J.; Murray, B.; et al. Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. J. Biol. Chem. 2001, 276, 19404–19413.
- Galindo-Trigo, S.; Grand, T.M.; Voigt, C.A.; Smith, L.M. A malectin domain kinesin functions in pollen and seed development in Arabidopsis. J. Exp. Bot. 2020, 71, 1828–1841.
- Gu, F.; Bringmann, M.; Combs, J.R.; Yang, J.; Bergmann, D.C.; Nielsen, E. Arabidopsis CSLD5 Functions in Cell Plate Formation in a Cell Cycle-Dependent Manner. Plant Cell 2016, 28, 1722–1737.
- Eggermont, L.; Verstraeten, B.; Van Damme, E.J.M. Genome-wide screening for lectin motifs in Arabidopsis thaliana. Plant Genome 2017, 10, 1–17.
- Slavokhotova, A.A.; Shelenkov, A.A.; Andreev, Y.A.; Odintsova, T.I. Hevein-like antimicrobial peptides of plants. Biochem. (Mosc.) 2017, 82, 1659–1674.
- Barad, S.; Sela, N.; Dubey, A.K.; Kumar, D.; Luria, N.; Ment, D.; Cohen, S.; Schaffer, A.; Prusky, D. Differential gene expression in tomato fruit and Colletotrichum gloeosporioides during colonization of the RNAi–SlPH tomato line with reduced fruit acidity and higher pH. BMC Genom. 2017, 18, 579.
- Xu, J.; Wang, X.Y.; Guo, W.Z. The cytochrome P450 superfamily: Key players in plant development and defense. J. Integrat. Agric. 2015, 14, 1673–1686.
- Bathe, U.; Tissier, A. Cytochrome P450 enzymes: A driving force of plant diterpene diversity. Phytochemistry 2019, 161, 149–162.
- Chen, Y.; Peumans, W.J.; Hause, B.; Bras, J.; Kumar, M.; Proost, P.; Barre, A.; Rouge, P.; Van Damme, E. Jasmonate methyl ester induces the synthesis of a cytoplasmic/nuclear chitooligosaccharide-binding lectin in tobacco leaves. FASEB J. 2002, 16, 905–907.
- Delporte, A.; De Zaeytijd, J.; De Storme, N.; Azmi, A.; Geelen, D.; Smagghe, G.; Guisez, Y.; Van Damme, E. Cell cycle-dependent O-GlcNAc modification of tobacco histones and their interaction with the tobacco lectin. Plant Physiol. Biochem. 2014, 83, 151–158.
- Cui, X.; Yan, Q.; Gan, S.; Xue, D.; Wang, H.; Xing, H.; Zhao, J.; Guo, N. GmWRKY40, a member of the WRKY transcription factor genes identified from Glycine max L., enhanced the resistance to Phytophthora sojae. BMC Plant Biol. 2019, 19, 598.
- Jiang, Y.; Yu, D. The WRKY57 Transcription Factor Affects the Expression of Jasmonate ZIM-Domain Genes Transcriptionally to Compromise Botrytis cinerea Resistance. Plant Physiol. 2016, 171, 2771–2782.
- Chen, L.; Zhang, L.; Xiang, S.; Chen, Y.; Zhang, H.; Yu, D. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 2021, 72, 1473–1489.
- Liu, J.; Chen, X.; Liang, X.; Zhou, X.; Yang, F.; Liu, J. Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol. 2016, 171, 1427–1442.
- Stefanowicz, K.; Lannoo, N.; Zhao, Y.; Eggermont, L.; Hove, J.V.; Atalah, B.A.; Van Damme, E.J.M. Glycan-binding F-box protein from Arabidopsis thaliana protects plants from Pseudomonas syringae infection. BMC Plant Biol. 2016, 16, 213.
- Zaltsman, A.; Krichevsky, A.; Loyter, A.; Citovsky, V. Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe 2010, 7, 197–209.
- Bishop, C.D.; Cooper, R.M. An ultrastructural study of vascular colonization in 3 vascular wilt diseases. 1. Colonization of susceptible cultivars. Physiol. Plant Pathol. 1983, 23, 323–343.
- Ndimba, B.K.; Chivasa, S.; Hamilton, J.M.; Simon, W.J.; Slabas, A.R. Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics 2003, 3, 1047–1059.
- Faruque, K.; Begam, R.; Deyholos, M.K. The amaranthin-like lectin (LuALL) genes of flax: A unique gene family with members inducible by defence hormones. Plant Mol. Biol. Rep. 2015, 33, 731–741.
- Cole, S.J.; Yoon, A.J.; Faull, K.F.; Diener, A.C. Host perception of jasmonates promotes infection by Fusarium oxysporum formae speciales that produce isoleucine- and leucine-conjugated jasmonates. Mol. Plant Pathol. 2014, 15, 589–600.
- Yamada, Y.; Nishida, S.; Shitan, N.; Sato, F. Genome-wide identification of AP2/ERF transcription factor-encoding genes in California poppy (Eschscholzia californica) and their expression profiles in response to methyl jasmonate. Sci. Rep. 2020, 10, 18066.
- Dang, L.; Rougé, P.; Van Damme, E.J.M. Amaranthin-like proteins with aerolysin domains in plants. Front Plant Sci. 2017, 8, 1368.
- Phillips, M.A.; Walter, M.H.; Ralph, S.G.; Dabrowska, P.; Luck, K.; Uros, E.M.; Boland, W.; Strack, D.; Rodriguez-Concepcion, M.; Bohlmann, J.; et al. Functional identification and differential expression of 1-deoxy-D-xylulose 5-phosphate synthase in induced terpenoid resin formation of Norway spruce (Picea abies). Plant Mol. Biol. 2007, 65, 243–257.
- Zulak, K.; Bohlmann, J. Terpenoid biosynthesis and specialized vascular cells of conifer defense. J. Integr. Plant Biol. 2010, 52, 86–97.
- Boba, A.; Kostyn, K.; Kozak, B.; Wojtasik, W.; Preisner, M.; Prescha, A.; Gola, E.; Lysh, D.; Dudek, B.; Szopa, J.; et al. Fusarium oxysporum infection activates the plastidial branch of the terpenoid biosynthesis pathway in flax, leading to increased ABA synthesis. Planta 2020, 251, 50.




