Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Zhifang Lu | + 1850 word(s) | 1850 | 2022-03-10 03:13:00 | | | |
| 2 | Catherine Yang | Meta information modification | 1850 | 2022-03-21 02:53:04 | | | | |
| 3 | Catherine Yang | + 3 word(s) | 1853 | 2022-03-21 04:55:50 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lu, Z. YdfD. Encyclopedia. Available online: https://encyclopedia.pub/entry/20756 (accessed on 07 February 2026).
Lu Z. YdfD. Encyclopedia. Available at: https://encyclopedia.pub/entry/20756. Accessed February 07, 2026.
Lu, Zhifang. "YdfD" Encyclopedia, https://encyclopedia.pub/entry/20756 (accessed February 07, 2026).
Lu, Z. (2022, March 19). YdfD. In Encyclopedia. https://encyclopedia.pub/entry/20756
Lu, Zhifang. "YdfD." Encyclopedia. Web. 19 March, 2022.
Copy Citation
ydfD is a lytic gene from the Qin cryptic prophage that encodes a 63-amino-acid protein, the ectopic expression of which in Escherichia coli can cause nearly complete cell lysis rapidly. The bacterial 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway is responsible for synthesizing the isoprenoids uniquely required for sustaining bacterial growth.
IspG
YdfD
1. Introduction
Cryptic prophages, also known as defective prophages, are segments of phage DNA integrated and maintained in bacterial chromosomes. It has been shown that bacterial genomes can harbor multiple cryptic prophages [1][2][3][4]. In fact, as much as 10–20% of bacterial chromosome segments are estimated to be prophage genes, and a growing body of evidence suggests that the proportion may be even greater [1][3]. For example, it was in the 1950s that the E. coli K-12 genomes were first found to carry phage genes [5] and now there are 10 known cryptic prophages residing in the chromosome of E. coli K-12 [4]. The expression of most of the prophage genes is usually suppressed under normal conditions, as certain proteins they encode are typically lytic and, therefore, toxic to the host [6]. However, there is evidence that some prophage genes can not only endow their hosts with traits and behaviors beneficial to surviving harsh environmental conditions but also expand the hosts’ genetic diversity, which plays a key role in microbial evolution [7][8][9][10]. Despite recent progress made in understanding cryptic prophages, the relationship between cryptic prophages and hosts is still largely elusive.
Investigation of cryptic prophages may provide hints to understand the process of phage infection, especially phage-induced cell lysis. As far as presently known, phages establish canonical lytic pathways to escape from hosts by two proteins, holin and endolysin [11][12][13]. Holins form holes in the cell membrane, and endolysins are a class of hydrolases that degrade bacterial cell wall. Therefore, holins are considered to control the access of endolysins to the cell periplasm. Apart from degradation of the cell wall, some phage genes encode proteins to inhibit the key enzyme to prevent cell wall biosynthesis. These proteins were dubbed “single-gene lysis” (Sgl) [11]. Many phages encode lytic protein. However, there are only three phage targets that have been proposed: (1) the Leviviridae Qβ uses the protein A2 to bind and inhibit MurA, which is the first step in bacterial cell wall biosynthesis [14]; (2) the protein E from phage ΦX174 inhibits peptidoglycan synthesis by interaction with MraY, which involves the peptidoglycan (PG) biosynthesis pathway [15]; and (3) LysM inhibits the translocation of the final lipid-linked PG precursor across the cytoplasmic membrane by interfering with MurJ [16]. Therefore, phages being natural antibiotics, exploring the lysis targets of phages is timely and of considerable importance.
Q-independent (Qin) prophage was the third prophage identified, found in E. coli K-12 [17]. As a part of Qin prophage, ydfD encodes a 63-amino-acid protein and ydfD is located downstream of dicB. Both ydfD and dicB are regulated by dicA [18]. Overexpression of ydfD induces cell lysis in E. coli, whereas lysis is prevented by coexpression with the cell division inhibitor dicB or sulA. It is now known that DicB as well as SulA prevent cell division by inhibiting FtsZ polymerization [19][20]. This indicates that the cell lysis caused by YdfD depends on functional cell division. Based on what is currently known, YdfD is speculated to promote cell lysis by targeting an unknown unique host protein, although the identity of the target protein of YdfD has not been established [21].
2. YdfD Is a Lysis Protein of the Qin Prophage
The number of prophages identified in bacterial genomes or plasmids has increased substantially in recent years [4][22][23], although the underlying functional mechanisms of their host interactions have remained unclear [24]. Nevertheless, prophages appear to play key roles in host metabolism. Cryptic prophages have been shown to help bacteria resist antibiotic stress [24] and dicB, an upstream gene of ydfD, suppresses cell division in E. coli once induced [25]. Prophages can also prevent different categories of phages from infecting the host. DicB specifically inhibited infection by λ and other phages that use ManYZ membrane proteins to prevent phage DNA entrance [26], provide population-level benefits by promoting biofilm formation, and influence the evolution of their hosts [7][8][9].
A previous study showed that overexpression of ydfD caused cell lysis of E. coli [21], although the molecular mechanism was not illustrated. In this study, the researchers attempted to characterize YdfD and its underlying mechanism of cell lysis activity. Herein, the researchers report that YdfD interacts with the essential MEP enzyme IspG. The MEP pathway synthesizes precursors of the cell wall and membrane, and a variety of inhibitors have already been designed and screened against it [27][28]. It is thus tempting to suggest that YdfD-mediated cell lysis may be caused by YdfD inhibition of MEP production. This phenomenon may be regarded as a strategy for the prophage precursor to escape from the host under adverse conditions. This is analogous to the lytic function of protein E from phage ΦX174, which inhibits peptidoglycan synthesis by the MraY enzyme [15].
YdfD contains two well-conserved cysteine residues among bacterial species [21], and IspG has three conserved cysteine residues involved in the binding of the iron–sulfur cluster, responsible for the electron transfer of the catalytic process, as shown in Figure 1. The catalytic activity was significantly reduced when any cysteine that involved the cluster was replaced by serine [29]. However, the YdfD mutants lacking cysteine residue(s) had no effect on its binding to IspG, suggesting that the YdfD–IspG interaction is not through a disulfide bond. As the [4Fe-4S]2+ cluster is indispensable for IspG catalytic activity, all experimental procedures need to be performed under strict anerobic conditions [30]. In addition, ME-cPP, the substrate of IspG, is not commercially available. It is thus difficult to assay the effects of YdfD on IspG enzyme kinetics.
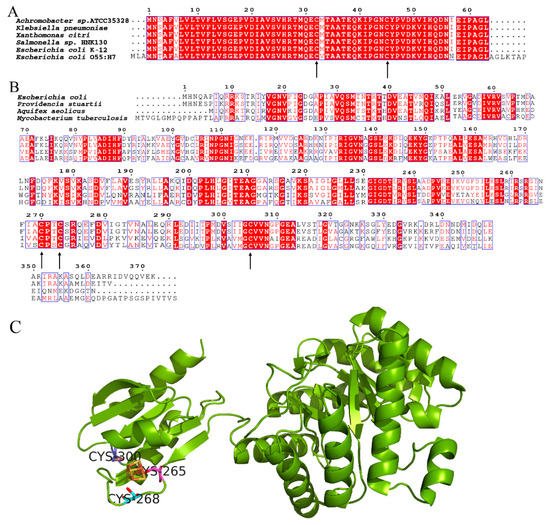
Figure 1. Conserved cysteine residues of YdfD and IspG. (A) Alignment of YdfD sequences from Achromobacter sp. ATCC35328, Klebsiella pneumoniae, Xanthomonas citri, Salmonella sp. , E. coli K-12, and E. coli O55:H7; the arrows indicate conserved cysteine residues. (B) Alignment of IspG sequences from E. coli, Providencia stuartii, Aquifex aeolicus, and Mycobacterium tuberculosis; the arrows indicate conserved cysteine residues. (C) Aquifex aeolicus IspG; three conserved cysteine residues binding the iron–sulfur cluster are responsible for the electron transfer of the catalytic process (PDB code: 3noy).
While it appears that the N-terminal domain of YdfD was responsible for binding to the N-terminal domain of IspG, intact YdfD was necessary for its full activity in the cell. It is possible that the N-terminal domain of YdfD recruits the C-terminal domain to IspG, which inhibits IspG’s activity, which is why both N-terminal and C-terminal domains of YdfD are essential for its cellular activity. This agrees with previous studies showing that the C-terminal domain of YdfD is essential for lysis but that the killing activity of the C-terminal domain alone is significantly reduced compared to that of the intact protein [21]. The Aquifex aeolicus IspG is known to fold into two domains: an N-terminal and a C-terminal domain [30]. In the Aquifex aeolicus IspG catalytic process, the substrate binds to the positively charged surface region at the N-terminal domain [31]. The ProtParam model (https://web.expasy.org/protparam/, accessed on 10 November 2021) [32] shows that the C-terminal domain of YdfD is negatively charged. Hence, it is possible that the C-terminal domain of YdfD competitively inhibits IspG by obstructing the binding of its substrate, ME-cPP, to the positively charged surface region of the IspG N-terminal domain. As a result, cooperation of both YdfD domains would be essential to inhibit IspG activity.
Under normal conditions, lysogenic phage levels remain steady. However, once the environment worsens, the transcription of lysis genes increases as the lysogenic phages look for ways to escape from the host [33][34][35]. Prophages also respond in this manner: although they cannot successfully assemble into viable phage particles, the lysis genes (ydfD) of Qin would increase transcription under adverse conditions. As expected, the RT-PCR of E. coli grown under stress conditions showed that mRNA levels of ydfD dramatically increases in non-acidic stress conditions and only slightly decreases under acidic conditions. The slight transcription decrease in the acidic medium is likely due to the bacteria’s general preference for more alkaline conditions and the corresponding reduction in the growth rate in a medium with a pH of 5.0 [36][37]. Consequently, the transcription of all unessential genes would be expected to decrease when bacteria are grown in an acidic environment.
The products of the MEP pathway are IPP and DMAPP, both of which are sole precursors for vital cellular components, including hopanoids [38]; menaquinone [39]; and polyprenyl phosphates, particularly undecaprenyl diphosphate (Und-p), a lipid carrier to synthesize peptidoglycan cell wall precursors that are essential to maintain cell integrity [40][41][42]. No matter how complex the terpenoid compounds are, all of them share a common five-carbon isoprene building block. A likely mechanism for YdfD-induced lysis is that YdfD blocks the MEP pathway in the cell membrane and cell wall biosynthesis. This would result in a shortage of key precursors for the cell wall and the cell membrane, leading to the abortion of cell wall formation, especially during cell division. A previous study by Masudo et al. found that YdfD cannot induce cell lysis when cell division is restrained [21]. The researchers suppose that YdfD cuts off the MEP pathway by interaction with IspG, resulting in deficient cell components and cell wall membrane. When E. coli cells divide normally, as cytomembrane and cell wall element synthesis is most intense at sites of division, YdfD blocks terpenoid synthesis, thus leading to cell lysis. However, when cell division is slowed or stalled, the need for terpenoids (e.g., Und-P) is reduced. Therefore, YdfD cannot lyse the nondividing cells, i.e., YdfD-mediated effects would depend on cell division [21]. Further studies are needed to explore the direct effects of YdfD on IspG. HMBPP synthesis is a committed step in the MEP pathway, and IspG is strongly conserved across bacterial species [43], so it is unsurprising that phages would adapt to target IspG specifically to disrupt the host physiology and exert their function. IspG will be the fourth identified target of the phage lysis proteins; the details of how YdfD binds IspG and interferes with HMBPP will likely require structure studies of the YdfD/IspG complex.
It is clear that DicB binds to MinC and prevents FtsZ ring formation. Moreover, DicB specifically inhibits infection by λ and other phages that use ManYZ membrane proteins to prevent phage DNA entrance [26]. dicB and ydfD are located in the same operon, with partial overlap. As mentioned before, DicB was able to offset YdfD-induced lysis [21]. The researchers propose the following model: in normal conditions, DicB inhibits cell division and prevents phage infection; when the environment gets worse, the transcription and expression of ydfD are activated, eventually leading to cell lysis (Figure 2).
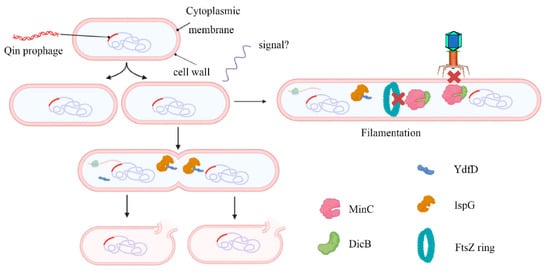
Figure 2. Working model for the regulation of host morphology by YdfD and DicB. The Qin cryptic prophage encodes the protein DicB and YdfD. When cell division is inhibited by DicB, demand for isoprenoid compounds is decreased and lysis is prevented (right). When YdfD is ectopically expressed, YdfD interacts with IspG, reducing isoprenoid production and causing the cells to lyse (left).
The growing severity of antibiotic resistance means there is a growing need to target novel bacterial pathways. The MEP pathway is absent in humans, making it an attractive target for antibiotic drug development [24]. The mechanisms used by prophages such as the Qin YdfD protein have proven to have a robust and widespread capacity to co-opt bacterial physiology. In addition to offering new insight into the particularities of host-prophage interactions, YdfD represents a promising new avenue for generating lead compounds for designing antibacterial agents.
References
- Arndt, D.; Marcu, A.; Liang, Y.J.; Wishart, D.S. PHAST, PHASTER and PHASTEST: Tools for finding prophage in bacterial genomes. Brief. Bioinform. 2019, 20, 1560–1567.
- Reis-Cunha, J.L.; Bartholomeu, D.C.; Manson, A.L.; Earl, A.M.; Cerqueira, G.C. ProphET, prophage estimation tool: A stand-alone prophage sequence prediction tool with self-updating reference database. PLoS ONE 2019, 14, e0223364.
- Song, W.C.; Sun, H.X.; Zhang, C.; Cheng, L.; Peng, Y.; Deng, Z.Q.; Wang, D.; Wang, Y.; Hu, M.; Liu, W.E.; et al. Prophage Hunter: An integrative hunting tool for active prophages. Nucleic Acids Res. 2019, 47, W74–W80.
- Blattner, F.R.; Plunkett, G.; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The complete genome sequence of Escherichia coli K-12. Science 1997, 277, 1453–1642.
- Lederberg, E.M.; Lederberg, J. Genetic Studies of Lysogenicity in Escherichia-Coli. Genetics 1953, 38, 51–64.
- Rueggeberg, K.G.; Toba, F.A.; Bird, J.G.; Franck, N.; Thompson, M.G.; Hay, A.G. The lysis cassette of DLP12 defective prophage is regulated by RpoE. Microbiology 2015, 161, 1683–1693.
- Stern, A.; Sorek, R. The phage-host arms race: Shaping the evolution of microbes. Bioessays 2011, 33, 43–51.
- Obeng, N.; Pratama, A.A.; van Elsas, J.D. The Significance of Mutualistic Phages for bacterial Ecology and Evolution. Trends Microbiol. 2016, 24, 440–449.
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327.
- Casjens, S. Prophages and bacterial genomics: What have we learned so far? Mol. Microbiol. 2003, 49, 277–300.
- Chamakura, K.; Young, R. Phage single-gene lysis: Finding the weak spot in the bacterial cell wall. J. Biol. Chem. 2019, 294, 3350–3358.
- Wang, I.N.; Smith, D.L.; Young, R. Holins: The protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000, 54, 799–825.
- Zampara, A.; Sørensen, M.C.H.; Grimon, D.; Antenucci, F.; Vitt, A.R.; Bortolaia, V.; Briers, Y.; Brøndsted, L. Exploiting phage receptor binding proteins to enable endolysins to kill Gram-negative bacteria. Sci. Rep. 2020, 10, 12087.
- Reed, C.A.; Langlais, C.; Kuznetsov, V.; Young, R. Inhibitory mechanism of the Qβ lysis protein A2. Mol. Microbiol. 2012, 86, 836–844.
- Bernhardt, T.G.; Struck, D.K.; Young, R. The lysis protein E of phi X174 is a specific inhibitor of the MraY-catalyzed step in peptidoglycan synthesis. J. Biol. Chem. 2001, 276, 6093–6097.
- Chamakura, K.R.; Sham, L.T.; Davis, R.M.; Min, L.; Cho, H.; Ruiz, N.; Bernhardt, T.G.; Young, R. A viral protein antibiotic inhibits lipid II flippase activity. Nat. Microbiol. 2017, 2, 1480–1484.
- Espion, D.; Kaiser, K.; Dambly-Chaudiere, C. A third defective lambdoid prophage of Escherichia coli K12 defined by the lambda derivative, lambdaqin111. J. Mol. Biol. 1983, 170, 611–633.
- Béjar, S.; Cam, K.; Bouché, J.P. Control of cell division in Escherichia coli. DNA sequence of dicA and of a second gene complementing mutation dicA1, dicC. Nucleic Acids Res. 1986, 14, 6821–6833.
- Chen, Y.; Milam, S.L.; Erickson, H.P. SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry 2012, 51, 3100–3109.
- Johnson, J.E.; Lackner, L.L.; de Boer, P.A. Targeting of (D)MinC/MinD and (D)MinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J. Bacteriol. 2002, 184, 2951–2962.
- Masuda, H.; Awano, N.; Inouye, M. ydfD encodes a novel lytic protein in Escherichia coli. FEMS Microbiol. Lett. 2016, 363, fnw039.
- Blaskovich, M.A.T.; Hansford, K.A.; Gong, Y.; Butler, M.S.; Muldoon, C.; Huang, J.X.; Ramu, S.; Silva, A.B.; Cheng, M.; Kavanagh, A.M.; et al. Protein-inspired antibiotics active against vancomycin- and daptomycin-resistant bacteria. Nat. Commun. 2018, 9, 22.
- Miura, K. An Overview of Current Methods to Confirm Protein-Protein Interactions. Protein Pept. Lett. 2018, 25, 728–733.
- Wang, X.X.; Kim, Y.; Ma, Q.; Hong, S.H.; Pokusaeva, K.; Sturino, J.M.; Wood, T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010, 1, 147.
- Cam, K.; Béjar, S.; Gil, D.; Bouché, J.P. Identification and sequence of gene dicB: Translation of the division inhibitor from an in-phase internal start. Nucleic Acids Res. 1988, 16, 6327–6338.
- Ragunathan, P.T.; Vanderpool, C.K. Cryptic-Prophage-Encoded Small Protein DicB Protects Escherichia coli from Phage Infection by Inhibiting Inner Membrane Receptor Proteins. J. Bacteriol. 2019, 201, e00475-19.
- Guerra, F.; Wang, K.; Li, J.K.; Wang, W.X.; Liu, Y.L.; Amin, S.; Oldfield, E. Inhibition of the 4Fe-4S proteins IspG and IspH: An EPR, ENDOR and HYSCORE investigation. Chem. Sci. 2014, 5, 1642–1649.
- Wang, W.; Li, J.; Wang, K.; Huang, C.; Zhang, Y.; Oldfield, E. Organometallic mechanism of action and inhibition of the 4Fe-4S isoprenoid biosynthesis protein GcpE (IspG). Proc. Natl. Acad. Sci. USA 2010, 107, 11189–11193.
- Zepeck, F.; Gräwert, T.; Kaiser, J.; Schramek, N.; Eisenreich, W.; Bacher, A.; Rohdich, F. Biosynthesis of Isoprenoids. Purification and Properties of IspG Protein from Escherichia coli. J. Org. Chem. 2005, 70, 9168–9174.
- Quitterer, F.; Frank, A.; Wang, K.; Rao, G.; O’Dowd, B.; Li, J.; Guerra, F.; Abdel-Azeim, S.; Bacher, A.; Eppinger, J.; et al. Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the Enzyme IspG (GcpE). J. Mol. Biol. 2015, 427, 2220–2228.
- Lee, M.; Gräwert, T.; Quitterer, F.; Rohdich, F.; Eppinger, J.; Eisenreich, W.; Bacher, A.; Groll, M. Biosynthesis of isoprenoids: Crystal structure of the cluster protein IspG. J. Mol. Biol. 2010, 404, 600–610.
- Gasteiger, E.H.C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607.
- Oppenheim, A.B.; Kobiler, O.; Stavans, J.; Court, D.L.; Adhya, S. Switches in bacteriophage lambda development. Annu. Rev. Genet. 2005, 39, 409–429.
- Erez, Z.; Steinberger-Levy, I.; Shamir, M.; Doron, S.; Stokar-Avihail, A.; Peleg, Y.; Melamed, S.; Leavitt, A.; Savidor, A.; Albeck, S.; et al. Communication between viruses guides lysis-lysogeny decisions. Nature 2017, 541, 488–493.
- Zong, C.H.; So, L.H.; Sepulveda, L.A.; Skinner, S.O.; Golding, I. Lysogen stability is determined by the frequency of activity bursts from the fate-determining gene. Mol. Syst. Biol. 2010, 6, 440.
- Hickey, E.W.; Hirshfield, I.N. Low-pH-induced effects on patterns of protein synthesis and on internal pH in Escherichia coli and Salmonella typhimurium. Appl. Environ. Microbiol. 1990, 56, 1038–1045.
- Booth, I.R. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 1985, 49, 359–378.
- Sandoval-Calderón, M.; Guan, Z.; Sohlenkamp, C. Knowns and unknowns of membrane lipid synthesis in streptomycetes. Biochimie 2017, 141, 21–29.
- Upadhyay, A.; Fontes, F.L.; Gonzalez-Juarrero, M.; McNeil, M.R.; Crans, D.C.; Jackson, M.; Crick, D.C. Partial Saturation of Menaquinone in Mycobacterium tuberculosis: Function and Essentiality of a Novel Reductase, MenJ. ACS Cent. Sci. 2015, 1, 292–302.
- Crick, D.C.; Schulbach, M.C.; Zink, E.E.; Macchia, M.; Barontini, S.; Besra, G.S.; Brennan, P.J. Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J. Bacteriol. 2000, 182, 5771–5778.
- Bongers, M.; Chrysanthopoulos, P.K.; Behrendorff, J.B.Y.H.; Hodson, M.P.; Vickers, C.E.; Nielsen, L.K. Systems analysis of methylerythritol-phosphate pathway flux in E. coli: Insights into the role of oxidative stress and the validity of lycopene as an isoprenoid reporter metabolite. Microb. Cell Factories 2015, 14, 193.
- Chang, S.-Y.; Ko, T.-P.; Liang, P.-H.; Wang, A.H.J. Catalytic Mechanism Revealed by the Crystal Structure of Undecaprenyl Pyrophosphate Synthase in Complex with Sulfate, Magnesium, and Triton. J. Biol. Chem. 2003, 278, 29298–29307.
- Wang, W.; Oldfield, E. Bioorganometallic chemistry with IspG and IspH: Structure, function, and inhibition of the proteins involved in isoprenoid biosynthesis. Angew. Chem. Int. Ed. 2014, 53, 4294–4310.
More
Information
Subjects:
Microbiology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.6K
Entry Collection:
Tight Junction and Its Proteins
Revisions:
3 times
(View History)
Update Date:
21 Mar 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




