| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Wutigri Nimlamool | + 2008 word(s) | 2008 | 2020-09-18 03:28:47 | | | |
| 2 | Bruce Ren | Meta information modification | 2008 | 2020-09-18 05:44:53 | | | | |
| 3 | Bruce Ren | -13 word(s) | 1995 | 2020-09-18 05:54:17 | | |
Video Upload Options
Ubiquilins or UBQLNs, members of the ubiquitin-like and ubiquitin-associated domain (UBL-UBA) protein family, serve as adaptors to coordinate the degradation of specific substrates via both proteasome and autophagy pathways. The UBQLN substrates reveal great diversity and impact a wide range of cellular functions. For decades, researchers have been attempting to uncover a puzzle and understand the role of UBQLNs in human cancers, particularly in the modulation of oncogene’s stability and nucleotide excision repair.
1. Introduction
Ubiquilins (UBQLNs) are essential factors for the maintenance of proteostasis in cells since they work as adaptors to deliver poly-ubiquitinated proteins to the proteasome [1][2] and participate in autophagosome formation [3][4][5], as well as in the endoplasmic reticulum (ER)-associated protein degradation pathway (ERAD) [6].
Human genome encodes four major UBQLN proteins (UBQLN1–4) that belong to the non-proteasomal UBL-UBA family by containing an ubiquitin-like (UBL) domain at the N-terminus and an ubiquitin-associated domain (UBA) at the C-terminus (Figure 1). UBQLN1, UBQLN2 and UBQLN4 are ubiquitously expressed, while UBQLN3 is exclusively expressed in the testis. The fifth UBQLN gene, called UBQLNL, was later detected in humans and still very poorly characterized [7].
Given the broad physiological implication of proteostasis pathways, any dysregulation of proteostasis is often involved in the development of multiple pathological conditions. Accumulated evidence links UBQLNs to neurodegenerative diseases such as Alzheimer’s disease (AD) or another form of dementia with locomotor dysfunctions [8][9] and other proteinopathies like amyotrophic lateral sclerosis (ALS) [10][11][12]. Moreover, increasing evidence suggests a role of UBQLNs in diverse types of cancers due to their activities in the modulation of important players of cell cycle, apoptosis, membrane receptors, DNA repairs, epithelial-mesenchymal transition (EMT) and miRNAs. Nevertheless, the mechanisms of how UBQLNs are involved in tumorigenesis and cancer progression are fragmented or contradictory, making it difficult to assess the contribution of such a family of proteins to the network of human cancers.
Figure 1. Structural organization of human ubiquilins (UBQLNs). The figure depicts the functional domains of major UBQLNs (UBQLN1-4) that have been functionally linked to human diseases. Structural analyses of the UBQLN proteins were performed using the integrated tool InterProScan (available online at https://www.ebi.ac.uk/interpro/). UBQLN1, UBQLN2 and UBQLN4 proteins have very similar UBA and UBL domains, encode structurally identical proteins containing four STI domains and are related by successive duplications [7]. In contrast, UBQLN3 carries a single STI domain and has been proposed to be more evolutionary divergent. UBL: Ubiquitin-like domain; STI: STress Inducible proteins or Hsp70–Hsp90 organizing protein; UBA: Ubiquitin associating domain.
2. UBQLN’s Genetic Variants in Cancers
At least six different genetic variants of UBQLN2 have been found to clearly correlate with ALS (OMIM entry 300264 at www.omim.org). Moreover, a homozygous nonsense mutation (c.976c>T) of UQBLN4 gene was recently discovered to cause a rare inherited disorder called ataxia-telangiectasia (A-T) [13].
To date, none of UBQLN’s genetic variants has been associated with cancers. Nonetheless, as discussed above, new studies have shown that an alteration of UBQLNs expression levels and/or a formation of proteinaceous UBQLNs-containing cytoplasmic aggregates are certain conditions that lead to abnormal cell growth and genome instability [13].
By utilizing an open platform that interactively explores multidimensional cancer genomics data sets in the context of clinical data and biologic pathways (https://www.cbioportal.org) [14][15], we were able to track several UBQLN single nucleotide variations (SNVs) and copy number variations (CNVs) that are associated with a large variety of human cancers, including breast, ovarian cancers and lung adenocarcinoma. For instance, we found that gene amplification is prominently found among UBQLN variations associated with cancers. Amplification of UBQLN4 is associated with 14.96% of breast cancer (446 samples out of 3116 samples from both the Cancer Genome Atlas (TCGA) and non-TCGA non-redundant studies). Similarly, amplification of UBQLN4 can also be identified in 10.12% of ovarian cancer patients (59 samples out of 583 samples from both TCGA and non-TCGA non-redundant studies) .
By accessing the cbioportal for UBQLN’s SNVs, the data have shown that some UBQLN’s polymorphisms segregate with the cancer samples by a significantly high allele frequency, especially in breast invasive carcinoma tumor. We identified UBQLN1 non-sense mutations in 31% (Q176*) and 53% (G499*) of variants reads from the TCGA and non-TCGA non-redundant breast and lung cancers studies, respectively. Moreover, three missense mutations in UBQLN2 (P440Q, H90N and G481V) and UBQLN3 (M546I, I247L and S294L) genes, show allele frequency higher than 50% in both TCGA and non-TCGA non-redundant studies of NSCLC.
On the basis of these findings, it would be of interest to run new studies in order to understand the possible relevance of such UBQLN variants in the onset of cancers or their progression. On the other hand, it is important to explore the reliability of UBQLNs as biomarkers of human cancers.
In this regard, some studies have already tested the feasibility to include UBQLNs as new biomarker panels for clinical and prognostic purposes in diverse human cancers. For example, Shimada and co-workers showed that UBQLN2 immunostaining can be a practical test for urine cytology allowing the prediction of tumor grade and stage [16]. Profiling the UBQLN1 together with other peptides in serum samples from lung adenocarcinoma was found to be able to predict cancer status with 85% sensitivity and 86% specificity [17][18]. Moreover, clinical and prognostic significance of UBQLN1 was more deeply investigated in breast and gastric cancers as well as in NSCLC and it revealed that high levels of UBQLN1 are often associated with high histological grade, invasion and lymph node metastasis. Therefore, high UBQLN1 expression is considered to be a worse prognostic factor for patient with gastric cancer [19] and a novel marker to predict poor prognosis in breast cancer [20].
3. Emerging Evidence of Ubiquilins/ncRNAs Axis in Cancer
MicroRNAs (miRNAs) are small non-coding RNAs (19–22 nucleotides in length) that are known to be fundamental regulators of eukaryotic gene expression. miRNAs contribute to the regulation of a variety of biological processes, such as cell cycle, differentiation, proliferation, apoptosis, autophagy, stress tolerance, energy metabolism and immune response [21][22][23][24]. Furthermore, it is known that miRNAs play a critical role in cancer pathogenesis and the dysregulation of miRNAs is a well-known feature of cancer.
The miR-200 family is involved in the self-renewal of cancer stem cells, EMT and chemo-sensitivity[25][26][27][28]. Interestingly, in three independent online databases (Targetscan, Pictar and miRbase) UBQLN1 is identified to be the target of miR-200c, a member of miR-200 family. Further in vivo experiments have proven that mir-200c is capable of binding to the 3’-UTR of UBQLN1, thus the overexpression of mir-200c in MDA-MB-231 and BT549 cells can lead to a strong reduction of UBQLN1 mRNA levels. Therefore, Sun and co-workers demonstrated that miR-200c inhibits autophagy and enhances radio-sensitivity in breast cancer cells by targeting UBQLN1 (Figure 2A).
Figure 2. UBQLNs and miRNAs networking in cancers. Different kinds of miRNAs affect UBQLN’s functions that are relevant to cancer. (A) miR-200 and (B) miR-155 negatively control UBQLN1 functions in breast cancer and lung adenocarcinoma, respectively. (C) miR-675 positively controls UBQLN1 in pancreatic cancer. (D) The negative control of UBQLN4 by miR-370 in hepatocellular carcinoma is shown.
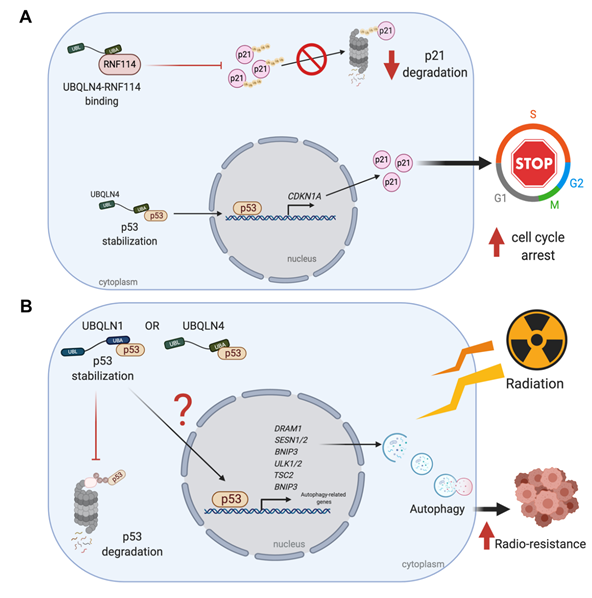
Similarly, through online prediction Yang and co-workers observed that miR-155 has two putative binding sites with UBQLN1 3’-UTR and further proved that overexpression of miR-155 reduces the expression of UBQLN1 mRNA and its protein in human nasopharyngeal carcinoma (NPC) cells [29]. Interestingly, miR-155 and antimir-155 showed abrogative and additive effects of UBQLN1, respectively; therefore, Yang and co-workers proposed that the miR-155/UBQLN1 axis is involved in multiple signaling pathways related to cell proliferation, cell cycle and EMT by regulating c-myc, E2F1 and cyclin D1. However, unlike the effects of miR-200c-dependent silencing of UBQLN1, the miR-155-dependent inhibition of UBQLN1 leads to an increase of radio-resistance in cancer cells because the overexpression of miR-155 abrogates the apoptosis inducing effects of UBQLN1 [29] (Figure 7B).
In line with the Yang and co-workers findings, other reports have shown that the Diesel exhaust Particles (DEP)-dependent downregulation of miR-155 leads to an augmentation of UBQLN1 and UBQLN2 mRNA levels in lung adenocarcinoma cell lines [30]. The study by Yadev and collaborators is the first to show an environmental carcinogen to regulate expression of UBQLN proteins and to suggest that UBQLNs are capable of protecting the cells from DEP toxicity. Interestingly, UBQLN1 proteins are frequently lost in lung cancer patients and the decreased UBQLN levels have proven to induce EMT in lung cancer cells (Figure 2B).
The miR-675 coding sequence lies within the first exon of the long-non-coding RNA (lncRNA) H19 [31]. miR-675 has been proven to cause reduction of cell proliferation, cell growth arrest and cell death and thus acts as a tumor suppressor in NSCLC and other cancer cells [32][33][34]. Interestingly, Wang and co-workers showed that miR-675 controls the abundance of UBQLN1 in a fashion that augmentation of miR-675 leads to an increase in the level of UBQLN1 which in turn reduces the level of ZEB1. Given the fundamental role of ZEB1 as a prime element of a network of transcription factors that controls EMT, the miR-675/UBQLN1 axis was shown to regulate the progression and development of pancreatic cancer by the regulation of ZEB1-dependent EMT [108] (Figure 2C).
The majority of studies, revealing the ncRNA/UBQLN axis in cancer, focus on the effects of miRNAs on UBQLN1. Recently, new findings suggested that such a fine cell regulation also involves other UBQLNs, like UBQLN4. Indeed, Yu and co-workers showed that miR-370/UBQLN4 axis regulates the formation and progression of hepatocellular carcinoma [35]. Given the emerging role of UBLQN4 in regulating cell proliferation and invasion [36], the online tools TargetScan, miRTarBase and miRcode were run to further investigate the upstream regulators of UBQLN4. In vivo studies showed that miR-370 binds to the UBQLN4 3’-UTR and leads to its degradation. Interestingly, the TCGA database analysis also confirmed that the expression of UBQLN4 negatively correlates with miR-370 expression [35]. As such, miR-370 seems to modulate the Wnt-β-catenin pathway that is controlled by UBQLN4 and is responsible for HCC progression (Figure 2D).
4. Conclusions and Future Perspectives
Although increasing evidence clearly points to the crucial roles of ubiqulin proteins in many cancers, recently there has been no ubiquilin-targeted therapy available in the market or in clinical trials. Parallel drugs that targets ubiquitin-protein degradation pathway including proteasome inhibitors (bortezomib, carfilzomib and ixazomib) are currently in clinical use. However, these three drugs which are mainly US FDA approved for relapse multiple myeloma and mantle cell lymphoma, provide decent treatment outcomes with many adverse effects. For instance, bortezomib can cause permanent nerve damage to the extremities, called bortezomib-induced peripheral neuropathy (BIPN). Furthermore, cancer cells developed resistance to this drug through an enhanced aggresome-autophagy pathway, increased expression of proinflammatory macrophages, decreased ER stress response and alterations in apoptotic signaling.
Considering the structure of UBQLNs, it contains similar domains at both N- and C-terminal regions to ubiquitin and it plays important roles in trafficking ubiquitinated proteins to the proteasome. Therefore, designing certain potential molecules to suppress the function of UBQLNs is not too difficult but the major challenge is to create the agents that can specifically target the mutated forms of UBQLNs in some cancer cells so that it does not affect normal healthy cells. Additionally, since UBQLNs play diverse functions in different types of cancer, these molecules can be “good” or “bad.” Therefore, developing only for ubiquilin inhibitors may not be a perfect solution for all type of cancers. Moreover, some cancer cells exhibit proliferative characteristics and tumorigenesis due to the lack of certain ubiquilin molecules, thus it needs to consider development of effective agents that can be able to restore the production of ubiquilin in a specific manner. Lastly, therapeutic strategies should be taken into consideration to optimize the best combinatorial treatments for patients. For instance, the inhibition of ubiquilin function together with radiation may enhance radiosensitivity.
Taken together, attempting to create personalized medicine for cancer patients with ubiquilin alterations may require an additional step to measure the level of certain ubiquilin, as well as its genetic and function abnormalities before choosing the most effective treatment for the patients.
References
- Seok Ko, H.; Uehara, T.; Tsuruma, K.; Nomura, Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004, 566, 110–114.
- Lee, D.Y.; Brown, E.J. Ubiquilins in the crosstalk among proteolytic pathways. Biol. Chem. 2012, 393, 441–447.
- N’Diaye, E.N.; Kajihara, K.K.; Hsieh, I.; Morisaki, H.; Debnath, J.; Brown, E.J. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009, 10, 173–179.
- Rothenberg, C.; Srinivasan, D.; Mah, L.; Kaushik, S.; Peterhoff, C.M.; Ugolino, J.; Fang, S.; Cuervo, A.M.; Nixon, R.A.; Monteiro, M.J. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum. Mol. Genet. 2010, 19, 3219–3232.
- Şentürk, M.; Lin, G.; Zuo, Z.; Mao, D.; Watson, E.; Mikos, A.G.; Bellen, H.J. Ubiquilins Regulate Autophagic Flux through mTOR Signaling and Lysosomal Acidification. Nat. Cell Biol. 2019, 21, 384.
- Lim, P.J.; Danner, R.; Liang, J.; Doong, H.; Harman, C.; Srinivasan, D.; Rothenberg, C.; Wang, H.; Ye, Y.; Fang, S.; et al. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J. Cell Biol. 2009, 187, 201–217.
- Marín, I. The ubiquilin gene family: Evolutionary patterns and functional insights. BMC Evol. Biol. 2014, 14, 63.
- Haapasalo, A.; Viswanathan, J.; Bertram, L.; Soininen, H.; Tanzi, R.E.; Hiltunen, M. Emerging role of Alzheimer’s disease-associated ubiquilin-1 in protein aggregation: Figure 1. Biochem. Soc. Trans. 2010, 38, 150–155.
- El Ayadi, A.; Stieren, E.S.; Barral, J.M.; Boehning, D. Ubiquilin-1 and protein quality control in Alzheimer disease. Prion 2013, 7, 164–169.
- Ling, S.-C.C.; Polymenidou, M.; Cleveland, D.W.W. Converging mechanisms in als and FTD: Disrupted RNA and protein homeostasis. Neuron 2013, 79, 416–438.
- Zhang, K.Y.; Yang, S.; Warraich, S.T.; Blair, I.P. Ubiquilin 2: A component of the ubiquitin-proteasome system with an emerging role in neurodegeneration. Int. J. Biochem. Cell Biol. 2014, 50, 123–126.
- Jantrapirom, S.; Lo Piccolo, L.; Yamaguchi, M. Non-Proteasomal UbL-UbA Family of Proteins in Neurodegeneration. Int. J. Mol. Sci. 2019, 20, 1893.
- Jachimowicz, R.D.; Beleggia, F.; Isensee, J.; Velpula, B.B.; Goergens, J.; Bustos, M.A.; Doll, M.A.; Shenoy, A.; Checa-Rodriguez, C.; Wiederstein, J.L.; et al. UBQLN4 Represses Homologous Recombination and Is Overexpressed in Aggressive Tumors. Cell 2019, 176, 505–519.
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal 2013, 6, pl1.
- Cerami1, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401.
- Shimada, K.; Fujii, T.; Tatsumi, Y.; Anai, S.; Fujimoto, K.; Konishi, N. Ubiquilin2 as a novel marker for detection of urothelial carcinoma cells in urine. Diagn. Cytopathol. 2016, 44, 3–9.
- Chen, G.; Wang, X.; Yu, J.; Varambally, S.; Yu, J.; Thomas, D.G.; Lin, M.-Y.; Vishnu, P.; Wang, Z.; Wang, R.; et al. Autoantibody Profiles Reveal Ubiquilin 1 as a Humoral Immune Response Target in Lung Adenocarcinoma. Cancer Res. 2007, 67, 3461–3467.
- Patel, K.; Farlow, E.C.; Kim, A.W.; Lee, B.S.; Basu, S.; Coon, J.S.; DeCresce, D.; Thimothy, L.; Walters, K.A.; Fhied, C.; et al. Enhancement of a multianalyte serum biomarker panel to identify lymph node metastases in non-small cell lung cancer with circulating autoantibody biomarkers. Int. J. Cancer 2011, 129, 133–142.
- Bao, J.; Jiang, X.; Zhu, X.; Dai, G.; Dou, R.; Liu, X.; Sheng, H.; Liang, Z.; Yu, H. Clinical significance of ubiquilin 1 in gastric cancer. Medicine 2018, 97, e9701.
- Wang, Y.; Lu, J.; Zhao, X.; Feng, Y.; Lv, S.; Mu, Y.; Wang, D.; Fu, H.; Chen, Y.; Li, Y. Prognostic significance of Ubiquilin1 expression in invasive breast cancer. Cancer Biomark. 2015, 15, 635–643.
- Wei, L.; Sun, J.; Zhang, N.; Zheng, Y.; Wang, X.; Lv, L.; Liu, J.; Xu, Y.; Shen, Y.; Yang, M. Noncoding RNAs in gastric cancer: Implications for drug resistance. Mol. Cancer 2020, 19, 62.
- Crudele, F.; Bianchi, N.; Reali, E.; Galasso, M.; Agnoletto, C.; Volinia, S. The network of non-coding RNAs and their molecular targets in breast cancer. Mol. Cancer 2020, 19, 61.
- Bhat, A.A.; Younes, S.N.; Raza, S.S.; Zarif, L.; Nisar, S.; Ahmed, I.; Mir, R.; Kumar, S.; Sharawat, S.K.; Hashem, S.; et al. Role of non-coding RNA networks in leukemia progression, metastasis and drug resistance. Mol. Cancer 2020, 19, 57.
- Almstrup, K.; Lobo, J.; Mørup, N.; Belge, G.; Rajpert-De Meyts, E.; Looijenga, L.H.J.; Dieckmann, K.-P. Application of miRNAs in the diagnosis and monitoring of testicular germ cell tumours. Nat. Rev. Urol. 2020, 17, 201–213.
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Downregulation of miRNA-200c Links Breast Cancer Stem Cells with Normal Stem Cells. Cell 2009, 138, 592–603.
- Jurmeister, S.; Baumann, M.; Balwierz, A.; Keklikoglou, I.; Ward, A.; Uhlmann, S.; Zhang, J.D.; Wiemann, S.; Sahin, O. MicroRNA-200c Represses Migration and Invasion of Breast Cancer Cells by Targeting Actin-Regulatory Proteins FHOD1 and PPM1F. Mol. Cell. Biol. 2012, 32, 633–651.
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601.
- Cochrane, D.R.; Howe, E.N.; Spoelstra, N.S.; Richer, J.K. Loss of miR-200c: A Marker of Aggressiveness and Chemoresistance in Female Reproductive Cancers. J. Oncol. 2010, 2010, 1–12.
- Yang, F.; Liu, Q.; Hu, C.M. Epstein-Barr virus-encoded LMP1 increases MIR-155 expression, which promotes radioresistance of nasopharyngeal carcinoma via suppressing UBQLN1. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 4507–4515.
- Yadav, S.; Singh, N.; Shah, P.P.; Rowbotham, D.A.; Malik, D.; Srivastav, A.; Shankar, J.; Lam, W.L.; Lockwood, W.W.; Beverly, L.J. MIR155 Regulation of Ubiquilin1 and Ubiquilin2: Implications in Cellular Protection and Tumorigenesis. Neoplasia 2017, 19, 321.
- Cai, X.; Cullen, B.R. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007, 13, 313.
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012, 14, 659–665.
- He, D.; Wang, J.; Zhang, C.; Shan, B.; Deng, X.; Li, B.; Zhou, Y.; Chen, W.; Hong, J.; Gao, Y.; et al. Down-regulation of miR-675-5p contributes to tumor progression and development by targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol. Cancer 2015, 14, 73.
- Wang, J.; Zhang, Y.; Wei, H.; Zhang, X.; Wu, Y.; Gong, A.; Xia, Y.; Wang, W.; Xu, M. The mir-675-5p regulates the progression and development of pancreatic cancer via the UBQLN1-ZEB1-mir200 axis. Oncotarget 2017, 8, 24978.
- Yu, Y.; Xu, P.; Cui, G.; Xu, X.; Li, K.; Chen, X.; Bao, J. UBQLN4 promotes progression of HCC via activating wnt-β-catenin pathway and is regulated by miR-370. Cancer Cell Int. 2020, 20, 3.
- Huang, S.; Dong, X.; Wang, J.; Ding, J.; Li, Y.; Li, D.; Lin, H.; Wang, W.; Zhao, M.; Chang, Q.; et al. Overexpression of the Ubiquilin-4 (UBQLN4) is Associated with Cell Cycle Arrest and Apoptosis in Human Normal Gastric Epithelial Cell Lines GES-1 Cells by Activation of the ERK Signaling Pathway. Med. Sci. Monit. 2018, 24, 3564.