Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mahsa Arabahmadi | + 5300 word(s) | 5300 | 2022-03-07 09:26:25 | | | |
| 2 | Vivi Li | + 46 word(s) | 5346 | 2022-03-16 10:35:49 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Arabahmadi, M. Deep Learning for Smart Healthcare. Encyclopedia. Available online: https://encyclopedia.pub/entry/20621 (accessed on 02 March 2026).
Arabahmadi M. Deep Learning for Smart Healthcare. Encyclopedia. Available at: https://encyclopedia.pub/entry/20621. Accessed March 02, 2026.
Arabahmadi, Mahsa. "Deep Learning for Smart Healthcare" Encyclopedia, https://encyclopedia.pub/entry/20621 (accessed March 02, 2026).
Arabahmadi, M. (2022, March 16). Deep Learning for Smart Healthcare. In Encyclopedia. https://encyclopedia.pub/entry/20621
Arabahmadi, Mahsa. "Deep Learning for Smart Healthcare." Encyclopedia. Web. 16 March, 2022.
Copy Citation
Advances in technology have been able to affect all aspects of human life. For example, the use of technology in medicine has made significant contributions to human society. Every year, many people die due to brain tumors; based on “braintumor” website estimation in the U.S., about 700,000 people have primary brain tumors, and about 85,000 people are added to this estimation every year. To solve this problem, artificial intelligence has come to the aid of medicine and humans. Magnetic resonance imaging (MRI) is the most common method to diagnose brain tumors. Additionally, MRI is commonly used in medical imaging and image processing to diagnose dissimilarity in different parts of the body.
smart healthcare
brain tumor classification
MRI
deep neural networks
CNN
GAN
transfer learning
1. Background
1.1. MRI Images—Segmentation and Classification
To segment, the region of interest (ROI) in image processing methods uses image segmentation and classification. In understanding images, extracting features, analyzing and interpreting images, image segmentation and classification have fundamental roles. A method to find the region of interest (ROI) or dividing the image into different regions or areas is medical image segmentation.
Failure to identify the exact location of the brain tumor leads to an incomplete, improper evacuation of the tumor, which causes the tumor to regrow and metastasize. These cases increase the risk of death. Image processing methods can be used to prevent and minimize this issue. For MR images, manual, semi-automatic or fully automatic techniques can be used. In medical image processing, manual techniques are time consuming and not as accurate as semi-automatic or fully automatic techniques. In addition, the design of a fully automated and effective classification still needs a second look because medical problems are related to human life and expert opinions are very decisive. Researchers have proposed several methods to develop such knowledge bases and, thus, the ability of tumor detection systems. MRI scanning is the most popular and general technique in neurology for imaging detailed specifications of the brain and other cranial structures. MRI can reveal flowing blood and hidden vascular malfunctions. The MRI scan is also beneficial and helpful for other brain-relevant diseases, such as Alzheimer’s disease [1], Parkinson’s disorder [2], and dementia [3]. The effect of COVID-19 on brain tissue was also investigated in MRI images in [4][5], along with many more diseases. There are various datasets available for training and testing purposes. In Table 1, the common datasets used in MR image segmentation are given.
Table 1. Available datasets of MR images.
| Dataset | Description | Ref. | Features |
|---|---|---|---|
| BRATS | Brain Tumor Segmentation Challenge (BRATS) always focus on the evaluation of current and novel methods for brain tumors segmentation in multimodal MR images and has the dataset available from 2012 to 2020. | [6][7][8] | Fully Convolutional Neural Network (FCNN) and Conditional Random Fields (CRF) used in Brain tumor segmentation and this is based in conjunction with the MICCAI 2012 and 2013 conferences. |
| OASIS | Open Access Series of Imaging Studies is contained over 2000 MR sessions are collected among several ongoing projects through the WUSTL Knight ADRC | [9][10] | Diagnosis of Alzheimer’s Disease. |
| TCIA | The Cancer Imaging Archive (TCIA) is a big archive of cancer images and available for public download. | [11][12][13] | Prediction of head and neck cancer and Prediction of pancreatic cancer. Segmentation of brain tumors. |
| IBSR | The Internet Brain Segmentation Repository. Its goal is to encourage the evaluation and expansion of segmentation methods. | [14][15][16] | Segmentation of MRI images and skull stripping. |
| BrainWeb | It is a Simulated Brain Database. | [17][18][19] | Reconstruction of 3D MR images based on CNN and reduction of noise from MRI images and segmentation of cerebrospinal fluid and brain volume-based CNN |
| NBIA | National Biomedical Imaging Archive that is for in vivo images, these images are related to biomedical research community, industry, and academia with access to image archives. | [20] | Quantitative Imaging Network. |
| The Whole Brain Atlas | This site has dozens of real images of the brain and the Harvard Whole Brain Atlas provides you with access to PET and MRI scans of normal and diseased brains. | [21][22] | Features Extracted from brain images by CNN and Serotonin Neurons. |
| ISLES | Ischemic Stroke Lesion Segmentation a medical image segmentation challenge at the MICCAI 2018 and a new dataset is consist of 103 stroke patients and matches profesional segmentations. | [23][24] | Brain lesion segmentation and stroke lesion segmentation. |
An automatic model can solve partially this problem, for instance, researchers can use the abnormality and object detection methods. The efficiency of automatic techniques belongs just to the knowledge databases in the absence of experts. Researchers have improved many methods to use automatic techniques and knowledge databases to improve the capability of tumor detection systems [25].
1.2. AI techniques
1.2.1. Deep Learning
Deep learning shows machine learning methods with multiple levels of representation. It consists of several layers, and the input of the layer is a representation from previous levels; with this structure, very complex features and inferences can be learned. A great deal of attention has been given to deep learning over the past several years for many applications in different areas, such as anomaly detection, image or object detection, pattern recognition, natural language processing. Deep learning can achieve great success in applications such as anomaly detection, image detection, pattern recognition, and natural language processing. Convolutional neural networks (CNNs), pre-trained unsupervised networks (PUNs), and recurrent/recursive neural networks (RNNs) are three different categories of deep learning.
Deep learning in the healthcare system equips doctors and experts to analyze any disease more accurately and helps them in implementing treatment and improve decision making. In Figure 1, some usages of deep learning in healthcare are shown [26][27][28][29][30][31][32][33][34][35].

Figure 1. Deep learning in healthcare.
1.2.2. Neural Network
Neural networks are chains of algorithms that mimic the operations of a human brain to recognize relationships between vast amounts of data. In recent years, DL and neural network (NN) can be superior to classical methods in object recognition methods. NNs can learn complex hierarchical representations of images because they have a strong representational power and thus, are more and more representing abstract concepts. They are strong at generalizing never-seen data. This feature empowers them to recognize a multitude of different objects whose appearance also varies greatly. For neuroscientists, a new approach for complex behaviors, heterogeneous neural activity in neural systems is provided by NNs. The benefit of neural networks is the facility of end-to-end training, and the action that neural networks generalize never seen data very powerful.
1.2.3. Machine Learning and Image Processing
Machine learning is a natural outgrowth of the intersection of computer science and statistics. The learning in machine learning happens via independent optimization of internal components, which are called parameters. Machine learning methods need careful engineering and expertise in domain knowledge to design feature extractors: So Yann LeCun designed and introduced a convolution neural network (CNN) that can learn to extract features automatically. The development of machine learning and soft computing approaches have created a remarkable impact in medical imaging too [36]. The efficiency of machine learning methods is related to the choice of data features on which they are applied.
The value of machine learning for radiomic features extraction in images is idiomatic, and was first introduced by Lambin in [37]. They described the solid cancer limitation, which gives huge potential for medical imaging and extracting features. Radiomics can extract wide amounts of image features from radiographic images and addresses this problem but needs better validation in multi-centric settings and the lab. Usually, radiomic features determine one scalar value to describe a whole three-dimensional (3D) tumor volume. Some features have connections with outcomes that can be fed into a classifier; the decision tree is an example of a useful classifier. ML defines the main aspects that generate the greatest predictive ability.
1.3. Deep Learning Applications
1.3.1. Anomaly Detection
In analyzing data, researchers can identify entities which are not similar to others, known as anomalies and also called outliers. Deep learning-based anomaly detection algorithms have become popular recently. In deep anomaly detection (DAD), learning methods are based on hierarchical discriminative features from data [38]. For both conventional and deep learning-based algorithms, a challenge is posed due to the absence of well-defined representative normal and anomalous border situations. Anomalies can be categorized as point anomalies, contextual anomalies, and collective anomalies. In anomaly detection, researchers have two types of data: sequential and non-sequential. Sequential data include video, speech, time series data, text (natural language); deep anomaly detection methods for these types are CNN, RNN, and LSTM. Non-sequential data include image, sensor, and other types of data; DAD methods for these types are CNN, AE and its variants. In addition, classification based on the type of deep learning models for anomaly detection can be (1) supervised, (2) semi-supervised, (3) unsupervised deep anomaly detection, (4) hybrid, and (5) one-class neural networks [38].
1.3.2. Object Detection
Object detection is mostly used in video analysis and image understanding. For the semantic perception of images and videos, object detection can present valuable information. Additionally, in fields such as image classification, human behavior analysis and face recognition, researchers can use object detection.
Generic object detection aims to locate and classify images and label them with rectangular bounding boxes to show the confidence of the entity [39]. So, CNN is one of the powerful ways to determine details. CNN is used in two aspects: CNN-based deep feature extraction and classification and localization. In object detection, CNN has some advantages compared to traditional methods, thus, these advantages, including video analysis, pedestrian detection, face recognition and image classification, are some examples of CNN research fields.
1.3.3. Pattern Recognition
In pattern recognition (PR), CNN has had significant success; for example, in an experimental realization of PR with CNN, for facial expression or emotion recognition, CNN design can be excellent. In the neuron model of PR, the multi-layer hierarchical network, there are forward and backward connections between cells, and CNN has good achievements. CNN is useful in text data, time-series data, and sequence input data. One of CNN’s abilities is to reduce data dimensionality, and the capability to classify in one network structure is a notable advantage of CNN.
1.3.4. Natural Language Processing
Natural language processing (NLP) can be named as one of the aspects of deep learning; In some cases, CNN is used in NLP, for example, in the case of utilizing CNNs for NLP inputs are sentences that they show as matrices. Each row of matrices are consist of a language element such as a words or a characters. To extract fixed features, CNN operation is good [40]. CNN achieves results in natural language processing applications because it can reduce most traditional problems. The convolutional architecture for language tasks is to apply a nonlinear (learned) function.
Natural language processing can be used for the classification of MRI reports. Unstructured text data, such as nursing records, reception reports, and discharge summaries, that are part of medical reports can be studied by NLP. NLP tools can be applied in a rule-based method to analyze the meaning of texts; moreover, several reports have used NLP to predict the progression of cancer or to classify breast pathology by analyzing free-text radiology reports [41][42].
2. Literature Review
2.1. Tumor Detection—Classic Approach
Segmentation, classification for tumor detection and localization based on MR images, is one of the concepts of medical image processing. Necessary specifications of brain tumor types and the identification of different segmentation and classification techniques that are successful for the detection of a range of brain diseases are presented in [25]. In this survey, the most relevant strategies, methods, working rules, preferences, constraints on MR images are covered. Designing an automatic algorithm to detect the brain tumor from MRI by artificial neural networks is studied in [43]; in this article set of image segmentation algorithms, feature extraction is proposed. The proposed algorithm was successfully tested and achieved the best results with an accuracy of 99% and sensitivity of 97.9%. A brain tumor detection system based on machine learning algorithms is proposed in [44], using gray level co-occurrence matrix (GLCM) to extract the texture-based features. In total, 212 samples of brain MR images are considered, and in the classification, perception and the Naive Bayes machine learning algorithm are used. The detection of tumors based on the programmed division strategy based on CNN is studied in [45]. For the detection of tumors from MRI images, the MATLAB tool is used by performing SVM classification. The processes and techniques used in detecting tumors based on medical imaging results are reviewed in [46]. A fully automatic brain tumor detection and segmentation method using the U-Net-based deep convolution networks is presented in [47]; they used BraTS 2015. The brain tumor segmentation model without manual interference is presented in this study. A mix of hand-crafted and deep learning features for segmentation image is presented in [48]; they used the grab–cut method for accurate segmentation. An automatic system for tumor extraction and classification from MRI based on marker-based watershed segmentation and features selection is developed in [49]. For diagnosis of the hardest brain tumor situation, in Ref, [50], the authors used deep CNN. They used MATLAB software, and their database consists of 1258 MRI images of 60 patients. The result of the study gained 96% accuracy.
2.2. Deep Learning and AI for Medical Imaging
AI methods for imaging methods have many suggestions to share image data better on different platforms [51]. Artificial intelligence improves radiologists’ diagnoses of malignant and benign tumors using MRI images in breast lesions [52]. AI allows radiologists to better diagnose, which may improve the patient’s therapy and cure. A high-performance algorithm to discover and characterize the presence of a meniscus tear on magnetic resonance imaging examination (MRI) of the knee is built in [53]. Analyzing the literature on artificial intelligence (AI) and radiomics, including all medical imaging modalities, for oncological and non-oncological applications for routine medical application is reviewed in [54]. A general outlook of deep learning-based MRI image processing and analysis is presented in [55]. A deep learning algorithm that can exactly diagnosis breast cancer on screening mammograms and uses an end-to-end training approach with a deep learning algorithm is developed in [56]. The main deep learning concepts which are related to medical image analysis are presented in [31].
2.3. CNN for Medical Imaging
Several tutorials and surveys have been published with a focus on convolutional neural networks and their challenges. Zhu Wenwu and his coworker in [57] in their book completely explained CNN structure with layers, and they explained CNN applications and architectures. They also described forwarding and backward propagation in CNN. Sergio Pereira et al. in [58], used normalization as a pre-processing step based on CNN. They worked on BraTS 2013 and 2015, and proposed an automatic segmentation method based on CNN. Darko Zikic et al. [59] studied the possibility to directly apply convolutional neural networks to the segmentation of brain tumor tissues. The matter is that the input to the network used multi-channel intensity information from a small patch around each point to be labeled. Only standard intensity pre-processing was applied to the input data to account for the scanner differences. No post-processing was applied to the output of the CNN. Jose Bernal et al. in their survey [60] presented a review of CNN techniques that are focused on architectures, pre-processing, data preparation and post-processing strategies in MRI images analysis. They reported how different CNN architectures have evolved, and also discussed state-of-the-art strategies.
Amin Kabir et al. in [61] used a method that derives from the combination of CNN and the genetic algorithm. They proposed to noninvasively classify different grades of glioma using MRI images to reduce the variance of prediction error, and utilized bagging as an ensemble algorithm. Jin Liu and Min Li [62] provided an overview of the concept of the brain tumor segmentation methods, object detection, registration and other tasks; the preprocessing methods for MRI-based brain tumor segmentation were introduced. A 3D fully connected conditional random field, which effectively removes false positives, was used in [23]. Additionally, for automatic lesion segmentation, they presented a 3D CNN architecture.
A dual-force training strategy was proposed in [63] to explicitly encourage deep models to learn high-quality multi-level features. The main point in [64] is a fully convolutional network whose input size is optional and generates a correspondingly sized output with effective inference and learning. Havaei et al. [65] proposed FCNN to segment and test images slice by slice. Additionally, a two-phase training scheme was proposed to deal with the class imbalance problem. Menz et al. [8] represented the multimodal brain tumor image segmentation. This method can be categorized as generative or discriminative. Fritscher et al. [66] presented a CNN for 3D-based deep learning components, which consists of three convolutional passes. A DCNN for multi-modal images was presented in [67]. Three architectures were proposed, whose patch (input) sizes are different. In addition, it proved that the size of the patch and the size of the convolutional filter affect the results when researchers use a patch way for segmenting brain tumors.
2.4. Modeling in CNN
The cascaded-CNN (C-CNN) is a novel deep learning architecture comprised of multiple CNNs [68]. CNN architectures are different in the depth of the network and the number of users. In continuation, three models based on the Multimodal Brain Tumor Segmentation Challenge (BraTS) are introduced.
2.4.1. Ensembles of Multiple Models and Architectures (EMMA)
This algorithm in the BraTS 2017 competition can obtain the first position between more than 50 teams. The performance of this algorithm is supreme because it combines multiple configured and trained CNN models [69]. Deep-Medic is the first employed architecture in this model; Deep-Medic is the 11-layers deep, multi-scale 3D CNN for brain lesion segmentation. EMMA can integrate two versions of 3D-Unet [69]. In the testing steps, any segmentation model segments images and outputs its class-confidence maps. EMMA is a deep learning model which can be run with great performance.
2.4.2. CNN-Based Segmentation of Medical Imaging Data
The performance of this model is similar to the U-Net CNN architecture with two rectifications: (1) merging multiple segmentation maps created at different scales, and (2) consigning feature maps from one stage of the network to another by using element-wise summation [69]. The CNN-based method with three-dimensional filters is demonstrated and applied to hand and brain MRI based on medical imaging data in [70]. In addition, two modifications to an existing CNN architecture are discussed, along with methods for addressing the aforementioned challenges. This model can achieve the first rank in BraTS 2015 and ischemic stroke lesion segmentation (ISLES).
2.4.3. Auto Encoder Regularization Based
This algorithm was introduced by Andriy Myronenko. This algorithm for extracting image features uses an encoder–decoder-based CNN architecture. For tumor subregion segmentation from 3D MRIs based on encoder–decoder architecture, a semantic segmentation network is used in [71]. The current approach won 1st place in the BraTS 2018 challenge.
2.5. Hybrid Techniques in Classification
Hybrid techniques integrate two or more techniques to obtain better results compared to individual techniques. In Table 2, some examples and usage of these techniques in analyzing MR images are given.
Table 2. Hybrid techniques in analyzing MR images.
| Technique | Ref. | Target | Result |
|---|---|---|---|
| Wavelet transform (WT), Genetic algorithm (GA) and supervised learning methods (SVM). | [72][73] | Classification of brain tissues in MRI images | This technique is accurate, Easy to operate, Non-invasive and inexpensive. |
| K-means, Sobel edge detection and morphological operations. | [74] | Segmentation of Brain Lesions in MRI and CT Scan Images | Achieves a high accuracy 94% in compared with manual delineation performed. |
| Support vector machine (SVM) and Fuzzy c-means (FCM). | [75][76] | Detection of Brain Tumor in MRI Images | Provide accurate and more effective result for classification of brain MRI images in minimal execution time. |
| K-Means, Nonsubsampled contourlet transform (NSCT) and SVM. | [77] | MRI Brain Tumor Images Classification | Higher classification accuracy. |
| K-Means, Gray Level Co-occurrence Matrix (GLCM), Berkeley Wavelet Transform (BWT), Principal Component Analysis (PCA) and Kernel Support Vector Machine (KSVM). | [78] | Detection and classification of MRI images | proposed method can be used for clinical purpose for screening and then diagnosed by the radiologists with high performance and accuracy. |
| Fuzzy Clustering, Gabor feature extraction and ANN. | [79] | Detection and Classification for Brain tumor | The classifier’s output helps the radiologist to make the decisions without any hesitation and achieved classification accuracy of 92.5%. |
3. Convolutional Neural Networks
This section focuses on the importance of CNN and the studies that are leveraging neural networks to tackle the problem of tumor detection. A comparison of different CNN architectures is reviewed in this section, and researchers talk about the usages of CNN methods in medicine.
3.1. Importance of CNN
Traditional neural networks are called the multilayer perceptron (MLP). MLP has drawbacks, such as using a perceptron for each input, which becomes uncontrollable for large-weight images. Another problem is the different response of MLP to an input (images) and its modified version. MLP cannot be a good option for image processing because spatial information is lost when the image is flattened into an MLP. One of the most effective methods of deep learning for image analysis to date, which has made noteworthy improvements in the image processing field, is convolutional neural networks. (CNNs/ConvNets) have many important achievements in resolving complex problems of machine learning. In neural networks, CNN is one of the main classes. CNN image classifications take input images, process these, and classify them with certain categories, e.g., cat and dog. The role of CNN is to decrease an image into a shape that is easier to process without losing any acute features, which are needed for a beneficent prediction. CNN has a powerful capability in processing images and learning features. CNN has a critical role in different deep neural network applications [57].
CNN has recently shown noted performance in computer vision for image segmentation and classification tasks; CNNs can learn the most useful features automatically. Convolutional layers that each input passes are kernel, pooling, fully connected, and SoftMax function. Figure 2 shows a complete CNN stream of processing an input image and classifying objects based on values. CNN contains many layers that transform their input with convolution filters of a small extent. Convolutional (sets of learnable filters), pooling (used to reduce overfitting and reduce image size) and fully connected (used to mix spatial and channel features together) are the three main layers of a convolutional network. The CNN layers are shown in Figure 2.
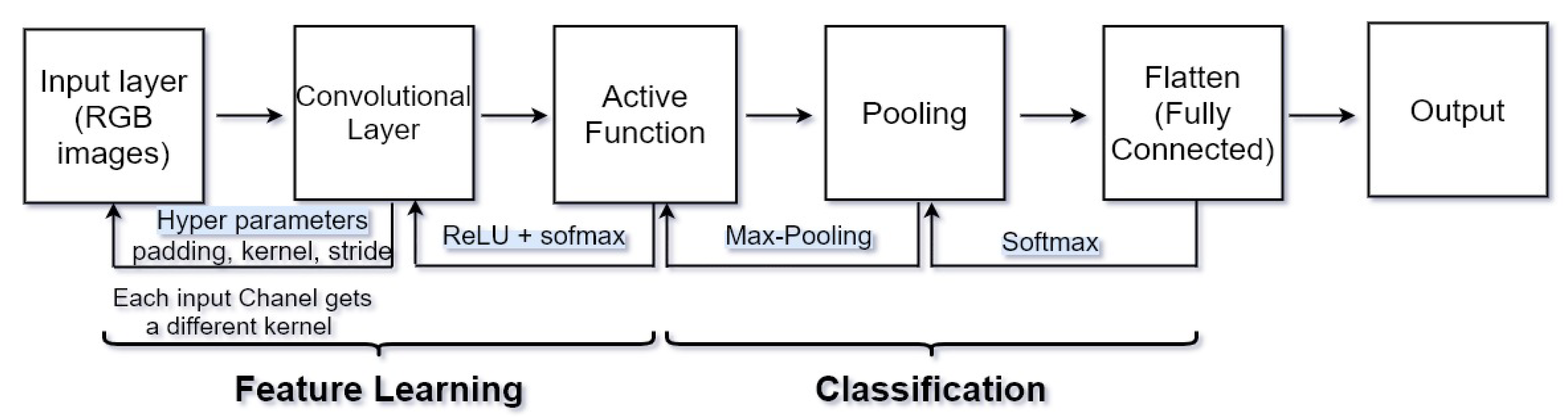
Figure 2. CNN layers, consist of 7 layers, input: [[CONV to RELU] × 2 to pool] × 3 to FC.
Convolution operators are used in most layers of these networks. In recent years, CNNs were applied in the segmentation of MS lesions, cerebral micro-bleeds and deep brain anatomical structures. High computational costs can be solved by convolutional networks, and this feature is very important since thousands of MRI images with different qualities and types are used for diagnosis, so CNN is used to classify brain tumor images. CNN can extract features automatically and further decrease the dimensions. CNN has performed well in processing medical images using deep neural networks. Deep learning algorithms, remarkably convolutional networks, have quickly become a methodology of election for analyzing medical images.
3.2. U-Net and Fully Convolutional Network
CNN has two main drawbacks: (1) the network works quite slowly because of the redundancy caused by the overlapping patches, and (2) the exchange that takes place between localization accuracy and classification accuracy [80]. Additionally, large patches use more context and they are more accurate, but they reduce the accuracy of the localization. For solving these problems, fully convolutional networks were introduced. Semantic information with appearance information are combined in FCN; FCN can produce accurate segmentation results [64]. Some good examples of FCN in achieving good results in medical image segmentation are shown in Refs. [81][82][83]. FCN is an instance of dense prediction networks.
U-shaped architecture, named U-Net, was developed by Ronneberger [84]. U-Net is a fully convolutional network that consists of a contracting path and an expansive path. The contracting path works as a feature extractor, and it follows the generic architecture of a convolutional network. The expansive path increases resolution by utilizing up-convolution; the ability of this network is obtaining the final segmentation results with only one training session [80]. U-Net gradually presents feature maps by connecting its encoding layers to its decoding layers of the same resolution for better perception and utilizes multi-level features [63]. Researchers can say that FCN and U-Net are dense prediction networks, and DeepMedic in another example of this network [23]. No pooling operations exist in DeepMedic, and the decrease in feature map size is realized by canceling padding operations in convolutional layers [63]. Researchers can say most of the current methods correlate multi-modality MRI data together as input. Table 3 provides a comparison of CNN modalities.
Table 3. Comparison of CNN modalities.
| Scheme | Dataset | Ref. | Ways of Training and Testing | Achievement |
|---|---|---|---|---|
| Rely on CNN | BRATS 2015 and ISLES 2015 | [23] | Dual pathway | An efficient solution processing for multi-scale processing for large image context using parallel convolutional pathways. |
| BRATS 2017 and BRATS 2015 | [63] | Dual-force | For learning high-quality multi-level features used a dual-force training strategy | |
| BRATS 2013 and BRATS 2015 | [58] | Patch-based | Used 3 × 3 kernels to permit deeper architectures for CNN-based segmentation method for brain MRI images. | |
| Rely on DCNN | ImageNet LSVRC-2010 | [85] | Patch-based | Gained top-1 and top-5 error rates of 37.5% and 17.0% |
| ISBI 2012& 2015 | [84] | End-to-end | Enabled precise localization. | |
| BRATS 2013 | [67] | T1, T1c, T2 and FLAIR images | 3D segmentation problem is converted into triplanar 2D CNNs. | |
| BRATS 2013 | [65] | T1, T1c, T2 and FLAIR images | Novel CNN architecture which improved accuracy and speed as presented in MICCAI 2013. | |
| Rely on FCN | BRATS 2013 & 2016 | [6] | T1, T1c, T2 and FLAIR images | Integration of FCN and Conditional Random Fields for brain tumor segmentation. |
| BRATS 2013 | [86] | End-to-end | Improve brain tumor segmentation performance by a symmetry-driven FCN | |
| ISBR and ABIDE (17 different sites) | [87] | End-to-end | Used 3D convolutional filters and FCN for an automatic segmentation of subcortical brain regions. |
3.3. Comparison of Different CNN Architectures
The most common CNN architectures are LeNet, AlexNet, VGGNet, GoogLeNet, ResNet, and ZFNet. They are implemented based on CNN. U-Net, SegNet and ResNet18 are the most popular CNNs for image segmentation [88].
LeNet, developed by Yann LeCun in the 1990s [89], is the first prosperous application of convolutional networks. Some usages of LeNet architecture are reading zip codes, digits, etc. One model of LeNet consists of five-layer CNN, which is called LeNet-5, and it can gain 99.2% accuracy on a single character recognition. The AlexNet, developed by Alex Krizhevsky [85], was the first convolutional network to become popular. The first five layers of AlexNet are convolutional layers, and the last three layers are fully connected layers, which contain eight main layers in total. To increase the speed and accuracy of AlexNet, ReLU is used. Microsoft Research in [90] proposed the residual neural network (ResNet). In ResNet, instead of unreferenced functions, layers are reformulated while learning residual functions. Residual networks have higher accuracy from increasing network depth, and the optimization of this network is easier. GoogLeNet was designed by Szegedy et al. [91], and it contains 22 layers. In comparison with AlexNet, it is much deeper. GoogLeNet contains 4 million parameters, and AlexNet contains 60 million parameters. One of the most used versions of GoogLeNet is Inception-v4. In Table 4, CNN architectures are compared with each other, and Table 5 provides some examples of CNN’s architectures in medicine.
Table 4. Comparison of different architecture of CNN.
| Ref. | Architectures | Layers | Advantages | Disadvantages |
|---|---|---|---|---|
| [92] | LeNet-5 | 7 layers | Ability to process higher resolution images need larger firmer layers. | Overfitting in some cases and no built-in mechanism to avoid this |
| [93] | AlexNet | 8 layers 60 M parameters | A very rapid downsampling of the intermediate representations through convolutions and max-pooling layers. | The use of large convolution filters (5 × 5) is not encouraged shortly after that, Is not deep enough rather than another techniques. |
| [94] | ZFNet | 8 layers | Improved image classification rate error in compared with Alexnet, winner of ILSVRC2012 | Feature maps are not divided across two different GPU, Thus connections between layers are dense. |
| [93] | GoogleNet | 22 layers 4–5 M parameters | Winner of ILSVRC2014, Decreased the number of parameters from 60 million (AlexNet) to 4 million so network can have a large width and depth. | Consists of a hierarchy of complex inception modules/blocks that consist of operations over different scales in each of the modules. |
| [95] | VGGNet | Between 11 to 19 layers the best one is 16 layers 138 M parameters | At present it is the most prefer election for extracting features from images. | Consists of 138 million parameters, which can be a bit challenging to handle. |
| [95] | ResNet | 152 layers | Network learns difference to an identity mapping (residual), Faster convergence if identity is closer to the optimum. | Lower complexity than VGGNet, Overfitting would increase test but decrease training error. |
Table 5. Architectures of CNN and their targets.
| Architectures | Examples | Target | Accuracy |
|---|---|---|---|
| LeNet-5 | [96] | Detection of brain cancer by tensorflow | 99% |
| [30] | classify Alzheimer’s brain | 96.85% | |
| Alex Net | [97] | Lung nodules in chest X-ray | 64.86% |
| [98] | Diagnosis of Thyroid Ultrasound Image | 90.8% | |
| [99] | Classification of skin lesion | 96.86% | |
| VGGNet-16 | [100] | Brain tumor classification | 84% |
| [101] | Diagnosis of Prostate Cancer | 95% | |
| Google Net | [102] | Thyroid Nodule Classification in Ultrasound Images | 98.29% |
| [97] | Lung nodules in chest X-ray | 68.92% | |
| ResNet | [103] | Brain tumor classification | 89.93% |
| [104] | Pancreatic tumor classification | 91% | |
| ZefNet | [105] | The trends and challenges for future edge reconfigurable platforms of deep learning. |
To purify the prediction outcomes of CNNs in network architectures, different post-processing methods were proposed. For instance, in [23], 3D-CRF was chosen for post-processing, which cures segmentation results by minimizing the Gibbs energy of every voxel. In addition, Havaei [65] presented clear predictions which are unusual in regions close to the skull according to the intensities of the voxels and the volume of the tumor area. A more complex post-processing pipeline is presented in [106], which is dependent on the voxel intensity, volume of the predicted area, etc. Setio et al. [107] used multi-view convolutional networks for pulmonary nodule detection; their network architecture is composed of multi-stream 2D CNNs.
3.4. Usages of CNN Methods in Medicine
In the U.S., breast cancer is the second major cause of cancer-related death. With mammography screening, mortality from breast cancer is reduced. The CAD system is used in mammography screening to improve the guessing accuracy. A modern CNN in the input has convolutional layers, and the output has one or more fully connected (FC) layers. In the paper of Shen et al. [56] was compared to methods of CNN, that are VGG and residual (Resnet) networks.
The visual geometry group (VGG) block is a stack of several 3 × 3 convolutional layers with 2 × 2 max pooling to reduce the feature map. The quality of the patch classifiers is important in the final classifiers’ results. Colorectal cancer (CRC) is the third most commonly diagnosed cancer [108]. To recognize and segment the exact location of tumors in CRC, MRI has good advantages. For the extraction of features from the colorectal tumor image, VGG-16 was used as the main model in [80], and for classification and localization information, five side-output blocks were used. In Table 6, the CNN methods in medicine are discussed.
Table 6. CNN methods in medical domain.
| Ref. | Features | Methods | Testing Sample | Achievement | Accuracy |
|---|---|---|---|---|---|
| [102] | Type, size, shape, tumor features | DCNN and googleNet | Thyroid nodules | Improving the performance of fine-tuning and augmenting the image samples. | 98.29% |
| [80] | Size, tumor features, doughnut-shaped lesion | FCN, VGG-16, U-Net | Colorectal tumors | Can remodel the current, time-consuming and non-reproducible manual segmentation method. | - |
| [104] | Type, size | ResNet18, ResNet34, ResNet52 and Inception-ResNet | Pancreatic Tumors | ResNet18 with the proposed weighted loss function method achieves the best results to classify tumors. | 91% |
| [107] | Type, size, shape | CAD system using a multi-view convolutional network | Pulmonary Nodule | Boosts the detection sensitivity from 85.7% to 93.3%. | - |
| [30] | Shape, scale | CNN and LeNet-5 | Alzheimer’s disease classification | Possible to generalize this method to predict different stages of Alzheimer’s disease for different age groups. | 96.85% |
| [99] | Type, color image lesions | transfer learning and Alex-net | skin lesions classification | Higher performance than existing methods. | 96.86% |
| [101] | Image lesion, type | VGGNet and patch-based DCNN | Prostate cancer | Enhanced prediction | 95% |
| [109] | Textures | AlexNet | Breast cancer | Showed that accuracy obtained by CNN on BreaKHis dataset was improved. | - |
Usages of CNN in E-Health
In recent years, CNN has been combined with the internet of things in wearable sensors toward improvement in the healthcare system. Progressive medical methods, such as telemedicine, image diagnosis, disease prediction, healthcare and so on, with the development of wearable sensors have been introduced. To mange daily life activities and healthcare, Ref. [110] focused on the wearable smart watch. In this paper, they used CNN for their target. A good example for the wearable sensor is presented in [111], a lightweight human bowel sounds (BSs) application.The recognizer is based on CNNs, and it is proposed for wearable systems. A lightweight CNN was used in [112] for the classification of a multivariate electroencephalogram (EEG). An online and accurate analysis for big data which are related to the brain is presented in this paper. Potentials of IoT technologies in brain healthcare are presented in [112]. Based on the collaborative machine learning approach in the field of IoT eHealth architecture, Ref. [113] reviewed arrhythmia detection by the use of CNN. The classification of tongue color based on CNN was studied in [114]; for training and testing images, they used CNN. Their experimental results showed that as the dataset increases, the accuracy becomes higher. For detecting tuberculosis in chest X-ray imaging, CNN was used in [115].
References
- Acharya, U.R.; Fernandes, S.L.; WeiKoh, J.E.; Ciaccio, E.J.; Fabell, M.K.M.; Tanik, U.J.; Rajinikanth, V.; Yeong, C.H. Automated Detection of Alzheimer’s Disease Using Brain MRI Images—A Study with Various Feature Extraction Techniques. J. Med. Syst. 2019, 43, 302.
- Amoroso, N.; Rocca, M.L.; Monaco, A.; Bellotti, R.; Tangaro, S. Complex networks reveal early MRI markers of Parkinson’s disease. Med. Image Anal. 2018, 48, 12–24.
- Bruun, M.; Koikkalainen, J.; Rhodius-Meester, E.A. Detecting frontotemporal dementia syndromes using MRI biomarkers. Neuroimage Clin. 2019, 22, 101711.
- Fitsiori, A.; Pugin, D.; Thieffry, C.; Lalive d’Epinay, P.; Vargas Gomez, M.I. Unusual Microbleeds in Brain MRI of Covid-19 Patients. J. Neuroimaging 2020, 30, 593–597.
- Espinosa, P.S.; Rizvi, Z.; Sharma, P.; Hindi, F.; Filatov, A. Neurological Complications of Coronavirus Disease (COVID-19): Encephalopathy, MRI Brain and Cerebrospinal Fluid Findings: Case 2. Cureus 2020, 12, e7930.
- Zhao, X.; Wu, Y. Brain Tumor Segmentation Using a Fully Convolutional Neural Network with Conditional Random Fields. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer International Publishing: Berlin/Heidelberg, Germany, 2016.
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, E. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. CoRR 2018, abs/1811.02629.
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024.
- Marcus, D.; Wang, T.; Parker, J.; Csernansky, J.; Morris, J.; Buckner, R. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. J. Cogn. Neurosci. 2007, 19, 1498–1507.
- Islam, J.; Zhang, Y. Early Diagnosis of Alzheimer’s Disease: A Neuroimaging Study With Deep Learning Architectures. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Salt Lake City, UT, USA, 18–22 June 2018.
- Diamant, A.; Chatterjee, A.; Vallières, M.; Shenouda, G.; Seuntjens, J. Deep learning in head & neck cancer outcome prediction. Sci. Rep. 2019, 9, 2764.
- Sekaran, K.; Chandana, P.; Krishna, N.M.; Kadry, S. Deep learning convolutional neural network (CNN) With Gaussian mixture model for predicting pancreatic cancer. Multimed. Tools Appl. 2020, 79, 10233–10247.
- AlBadawy, E.A.; Saha, A.; Mazurowski, M.A. Deep learning for segmentation of brain tumors: Impact of cross-institutional training and testing. Med. Phys. 2018, 45, 1150–1158.
- Kleesiek, J.; Urban, G.; Hubert, A.; Schwarz, D.; Maier-Hein, K.; Bendszus, M.; Biller, A. Deep MRI brain extraction: A 3D convolutional neural network for skull stripping. NeuroImage 2016, 129, 460–469.
- Pereira, S.; Pinto, A.; Oliveira, J.; Mendrik, A.M.; Correia, J.H.; Silva, C.A. Automatic brain tissue segmentation in MR images using Random Forests and Conditional Random Fields. J. Neurosci. Methods 2016, 270, 111–123.
- Shakeri, M.; Tsogkas, S.; Ferrante, E.; Lippe, S.; Kadoury, S.; Paragios, N.; Kokkinos, I. Sub-cortical brain structure segmentation using F-CNN’S. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 269–272.
- Jurek, J.; Kociński, M.; Materka, A.; Elgalal, M.; Majos, A. CNN-based superresolution reconstruction of 3D MR images using thick-slice scans. Biocybern. Biomed. Eng. 2020, 40, 111–125.
- Tripathi, P.C.; Bag, S. CNN-DMRI: A Convolutional Neural Network for Denoising of Magnetic Resonance Images. Pattern Recognit. Lett. 2020, 135, 57–63.
- Grimm, F.; Edl, F.; Kerscher, S.R.; Nieselt, K.; Gugel, I.; Schuhmann, M.U. Semantic segmentation of cerebrospinal fluid and brain volume with a convolutional neural network in pediatric hydrocephalus—Transfer learning from existing algorithms. Acta Neurochir. 2020, 162, 2463–2474.
- Kalpathy-Cramer, J.; Freymann, J.B.; Kirby, J.S.; Kinahan, P.E.; Prior, F.W. Quantitative Imaging Network: Data Sharing and Competitive AlgorithmValidation Leveraging The Cancer Imaging Archive. Transl. Oncol. 2014, 1, 147–152.
- Basheera, S.; Ram, M.S.S. Classification of Brain Tumors Using Deep Features Extracted Using CNN. J. Phys. Conf. Ser. 2019, 1172, 012016.
- Pollak Dorocic, I.; Fürth, D.; Xuan, Y.; Johansson, Y.; Pozzi, L.; Silberberg, G.; Carlén, M.; Meletis, K. A Whole-Brain Atlas of Inputs to Serotonergic Neurons of the Dorsal and Median Raphe Nuclei. Neuron 2014, 83, 663–678.
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.J.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78.
- Guerrero, R.; Qin, C.; Oktay, O.; Bowles, C.; Chen, L.; Joules, R.; Wolz, R.; Valdés-Hernández, M.D.; Dickie, D.A.; Wardlaw, J.; et al. White matter hyperintensity and stroke lesion segmentation and differentiation using convolutional neural networks. Neuroimage Clin. 2018, 43, 929–939.
- Chahal, P.K.; Pandey, S.; Goel, S. A survey on brain tumor detection techniques for MR images. Multimed. Tools Appl. 2020, 79, 21771–21814.
- Joudaki, H.; Rashidian, A.; Minaei-Bidgoli, B.; Mahmoodi, M.; Geraili, B.; Nasiri, M.; Arab, M. UsingnData Mining to Detect Health Care Fraud and Abuse: A Review of Literature. Glob. J. Health Sci. 2015, 7, 194–202.
- Roy, R.; George, K.T. Detecting insurance claims fraud using machine learning techniques. In Proceedings of the 2017 International Conference on Circuit, Power and Computing Technologies (ICCPCT), Kollam, India, 20–21 April 2017; pp. 1–6.
- Khagi, B.; Lee, C.G.; Kwon, G.R. Alzheimer’s disease Classification from Brain MRI based on transfer learning from CNN. In Proceedings of the 2018 11th Biomedical Engineering International Conference (BMEiCON), Chaing Mai, Thailand, 21–24 November 2018; pp. 1–4.
- Khedher, L.; Illan, I.A. Independent Component Analysis-Support Vector Machine-Based Computer-Aided Diagnosis System for Alzheimer’s with Visual Support. Int. J. Neural Syst. 2016, 27, 1650050.
- Sarraf, S.; Tofighi, G. Classification of Alzheimer’s Disease using fMRI Data and Deep Learning Convolutional Neural Networks. Sci. Rep. 2019, 9, 18150.
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88.
- Lavecchia, A. Deep learning in drug discovery: Opportunities, challenges and future prospects. Drug Discov Today 2019, 24, 2017–2032.
- Chen, H.; Engkvist, O.; Wang, Y.; Olivecrona, M.; Blaschke, T. The rise of deep learning in drug discovery. Drug Discov. Today 2018, 23, 1241–1250.
- Zampieri, G.; Vijayakumar, S.; Yaneske, E.; Angione, C. Machine and deep learning meet genome-scale metabolic modeling. PLoS Comput. Biol. 2019, 15, e1007084.
- Rusk, N. Deep learning. Nat. Methods 2016, 13, 35.
- Kamboj, A.; Rani, R.; Chaudhary, J. Deep Leaming Approaches for Brain Tumor Segmentation: A Review. In Proceedings of the 2018 First International Conference on Secure Cyber Computing and Communication (ICSCCC), Jalandhar, India, 15–17 December 2018; pp. 599–603.
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446.
- Chalapathy, R.; Chawla, S. Deep Learning for Anomaly Detection: A Survey. arXiv 2019, arXiv:1901.03407.
- Zhao, Z.; Zheng, P.; Xu, S.; Wu, X. Object Detection With Deep Learning: A Review. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 3212–3232.
- Yin, W.; Kann, K.; Yu, M.; Schutze, H. Comparative Study of CNN and RNN for Natural Language Processing. arXiv 2017, arXiv:1702.01923.
- Chen, P.H.; Zafar, H.; Galperin-Aizenberg, M.; Cook, T. Integrating Natural Language Processing and Machine Learning Algorithms to Categorize Oncologic Response in Radiology Reports. J. Digit. Imaging 2018, 31, 178–184.
- Yala, A.; Barzilay, R.; Salama, L.; Griffin, M.; Sollender, G.; Bardia, A.; Lehman, C.; Buckley, J.M.; Coopey, S.B.; Polubriaginof, F.; et al. Using machine learning to parse breast pathology reports. Breast Cancer Res. Treat. 2017, 161, 203–211.
- Abdalla, H.E.M.; Esmail, M. Brain tumor detection by using artificial neural network. In Proceedings of the 2018 International Conference on Computer, Control, Electrical, and Electronics Engineering (ICCCEEE), Khartoum, Sudan, 12–14 August 2018; pp. 1–6.
- Sharma, K.; Kaur, A.; Gujral, S. Brain Tumor Detection based on Machine Learning Algorithms. Int. J. Comput. Appl. 2014, 103, 7–11.
- Vinoth, R.; Venkatesh, C. Segmentation and Detection of Tumor in MRI images Using CNN and SVM Classification. In Proceedings of the Conference on Emerging Devices and Smart Systems (ICEDSS), Tiruchengode, India, 2–3 March 2018.
- Azhari, E.E.M.; Hatta, M.M.M.; Htike, Z.Z.; Win, S.L. Tumor Detection In Medical Imaging A Survey. Int. J. Adv. Inf. Technol. 2014, 4, 9.
- Dong, H.; Yang, G.; Liu, F.; Mo, Y.; Guo, Y. Automatic Brain Tumor Detection and Segmentation Using U-Net Based Fully Convolutional Networks. In Medical Image Understanding and Analysis; Valdés Hernández, M., González-Castro, V., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 506–517.
- Saba, T.; Sameh Mohamed, A.; El-Affendi, M.; Amin, J.; Sharif, M. Brain tumor detection using fusion of hand crafted and deep learning features. Cogn. Syst. Res. 2020, 59, 221–230.
- Khan, M.A.; Lali, I.U.; Rehman, A.; Ishaq, M.; Sharif, M.; Saba, T.; Zahoor, S.; Akram, T. Brain tumor detection and classification: A framework of marker-based watershed algorithm and multilevel priority features selection. Microsc. Res. Tech. 2019, 86, 909–922.
- Ahmed, B.; Al-Ani, M. An efficient approach to diagnose brain tumors through deep CNN. Math. Biosci. Eng. 2021, 18, 851–867.
- Thrall, J.H.; Li, X.; Li, Q.; Cruz, C.; Do, S.; Dreyer, K.; Brink, J. Artificial Intelligence and Machine Learning in Radiology: Opportunities, Challenges, Pitfalls, and Criteria for Success. J. Am. Coll. Radiol. 2018, 15, 504–508.
- Jiang, Y.; Edwards, A.V.; Newstead, G.M. Artificial Intelligence Applied to Breast MRI for Improved Diagnosis. Radiology 2020, 298, 38–46.
- Roblot, V.; Giret, Y.; Bou Antoun, M.; Morillot, C.; Chassin, X.; Cotten, A.; Zerbib, J.; Fournier, L. Artificial intelligence to diagnose meniscus tears on MRI. Diagn. Interv. Imaging 2019, 100, 243–249.
- Sollini, M.; Antunovic, L.; Chiti, A.; Kirienko, M. Towards clinical application of image mining: A systematic review on artificial intelligence and radiomics. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2656–2672.
- Liu, J.; Pan, Y.; Li, M.; Chen, Z.; Tang, L.; Lu, C.; Wang, J. Applications of deep learning to MRI images: A survey. Big Data Min. Anal. 2018, 1, 1–18.
- Shen, L.; Margolies, L.R.; Rothstein, J.H.; Fluder, E.; McBride, R.; Sieh, W. Deep Learning to Improve Breast Cancer Detection on Screening Mammography. Sci. Rep. 2019, 9, 12495.
- Zhu, W.; Wang, X.; Cui, P. Deep Learning for Learning Graph Representations. In Deep Learning: Concepts and Architectures; Pedrycz, W., Chen, S.M., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 169–210.
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans. Med. Imaging 2016, 1240–1251.
- Zikic, D.; Ioannou, Y.; Brown, M.; Criminisi, A. Segmentation of Brain Tumor Tissues with Convolutional Neural Networks. In Proceedings of the MICCAI Workshop on Multimodal Brain Tumor Segmentation Challenge (BRATS), Boston, MA, USA, 14 September 2014.
- Bernal, J.; Kushibar, K.; Asfaw, D.S.; Valverde, S.; Oliver, A.; Marti, R.; Llado, X. Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: A review. arXiv 2017, arXiv:1712.03747.
- Kabir Anaraki, A.; Ayati, M.; Kazemi, F. Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern. Biomed. Eng. 2019, 39, 63–74.
- Liu, T.; Li, M.; Wang, J.; Wu, F.; Liu, T.; Pan, Y. A survey of MRI-based brain tumor segmentation methods. Tsinghua Sci. Technol. 2014, 19, 578–595.
- Chen, S.; Ding, C.; Liu, M. Dual-force convolutional neural networks for accurate brain tumor segmentation. Pattern Recognit. 2019, 88, 90–100.
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 12 June 2015.
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31.
- Fritscher, K.; Raudaschl, P.; Zaffino, P.; Spadea, M.F.; Sharp, G.C.; Schubert, R. Deep Neural Networks for Fast Segmentation of 3D Medical Images. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI, München, Germany, 17–21 October 2016; pp. 158–165.
- Zhao, L.; Jia, K. Deep Feature Learning with Discrimination Mechanism for Brain Tumor Segmentation and Diagnosis. In Proceedings of the 2015 International Conference on Intelligent Information Hiding and Multimedia Signal Processing (IIH-MSP), Adelaide, Australia, 23–25 September 2015; pp. 306–309.
- Moritz, S.A.; Pfab, J.; Wu, T.; Hou, J.; Cheng, J.; Cao, R.; Wang, L.; Si, D. Cascaded-CNN: Deep Learning to Predict Protein Backbone Structure from High-Resolution Cryo-EM Density Maps. Sci. Rep. 2019, 10, 572990.
- Moujahid, H.; Cherradi, B.; Bahatti, L. Convolutional Neural Networks for Multimodal Brain MRI Images Segmentation: A Comparative Study. In Smart Applications and Data Analysis; Hamlich, M., Bellatreche, L., Mondal, A., Ordonez, C., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 329–338.
- Kayalibay, B.; Jensen, G.; van der Smagt, P. CNN-based Segmentation of Medical Imaging Data. arXiv 2017, arXiv:1701.03056.
- Myronenko, A. 3D MRI Brain Tumor Segmentation Using Autoencoder Regularization. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; pp. 311–320.
- Kharrat, A.; Gasmi, K.; Messaoud, M.B.; Benamrane, N.; Abid, M. A Hybrid Approach for Automatic Classification of Brain MRI Using Genetic Algorithm and Support Vector Machine. Leonardo J. Sci. 2010, 17, 71–82.
- Kumar, S.; Dabas, C.; Godara, S. Classification of Brain MRI Tumor Images: A Hybrid Approach. Procedia Comput. Sci. 2017, 122, 510–517.
- Agrawal, R.; Sharma, M.; Singh, B.K. Segmentation of Brain Lesions in MRI and CT Scan Images: A Hybrid Approach Using k-Means Clustering and Image Morphology. J. Inst. Eng. Ser. 2018, 99, 173–180.
- Parveen; Singh, A. Detection of brain tumor in MRI images, using combination of fuzzy c-means and SVM. In Proceedings of the 2015 2nd International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 19–20 February 2015; pp. 98–102.
- Abdel-Maksoud, E.; Elmogy, M.; Al-Awadi, R. Brain tumor segmentation based on a hybrid clustering technique. Egypt. Inform. J. 2015, 16, 71–81.
- Saha, C.; Hossain, M.F. MRI brain tumor images classification using K-means clustering, NSCT and SVM. In Proceedings of the 2017 4th IEEE Uttar Pradesh Section International Conference on Electrical, Computer and Electronics (UPCON), Mathura, India, 26–28 October 2017; pp. 329–333.
- Islam, A.; Hossain, M.F.; Saha, C. A new hybrid approach for brain tumor classification using BWT-KSVM. In Proceedings of the 2017 4th International Conference on Advances in Electrical Engineering (ICAEE), Dhaka, Bangladesh, 28–30 September 2017; pp. 241–246.
- Virupakshappa, D.B.A. Computer Based Diagnosis System for Tumor Detection & Classification: A Hybrid Approach. Int. J. Pure Appl. Math. 2018, 118, 33–43.
- Jian, J.; Xiong, F.; Xia, W.; Zhang, R.; Gu, J.; Wu, X.; Meng, X.; Gao, X. Fully convolutional networks (FCNs)-based segmentation method for colorectal tumors on T2-weighted magnetic resonance images. Australas. Phys. Eng. Sci. Med. 2018, 41, 393–401.
- Luo, Y.; Cheng, H.; Yang, L. Size-Invariant Fully Convolutional Neural Network for vessel segmentation of digital retinal images. In Proceedings of the 2016 Asia-Pacific Signal and Information Processing Association Annual Summit and Conference (APSIPA), Jeju, Korea, 13–16 December 2016; pp. 1–7.
- Fu, H.; Xu, Y.; Wong, D.W.K.; Liu, J. Retinal vessel segmentation via deep learning network and fully-connected conditional random fields. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 698–701.
- Huang, L.; Xia, W.; Zhang, B.; Qiu, B.; Gao, X. MSFCN-multiple supervised fully convolutional networks for the osteosarcoma segmentation of CT images. Comput. Methods Programs Biomed. 2017, 143, 67–74.
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 234–241.
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Advances in Neural Information Processing Systems 25; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Monett, MO, USA, 2012; pp. 1097–1105.
- Shen, H.; Zhang, J.; Zheng, W. Efficient symmetry-driven fully convolutional network for multimodal brain tumor segmentation. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; pp. 3864–3868.
- Dolz, J.; Desrosiers, C.; Ben Ayed, I. 3D fully convolutional networks for subcortical segmentation in MRI: A large-scale study. NeuroImage 2018, 170, 456–470.
- Daimary, D.; Bora, M.B.; Amitab, K.; Kandar, D. Brain Tumor Segmentation from MRI Images using Hybrid Convolutional Neural Networks. Procedia Comput. Sci. 2020, 167, 2419–2428.
- LeCun, Y.; Boser, B.E.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.E.; Jackel, L.D. Handwritten Digit Recognition with a Back-Propagation Network. In Advances in Neural Information Processing Systems 2; Touretzky, D.S., Ed.; Morgan-Kaufmann: Burlington, MA, USA, 1990; pp. 396–404.
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. CVPR 2016, 770–778.
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper With Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2015.
- El-Sawy, A.; EL-Bakry, H.; Loey, M. CNN for Handwritten Arabic Digits Recognition Based on LeNet-5. In Proceedings of the International Conference on Advanced Intelligent Systems and Informatics, Cairo, Egypt, 9–11 September 2017; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 566–575.
- Grm, K.; Struc, V.; Artiges, A.; Caron, M.; Ekenel, H.K. Strengths and Weaknesses of Deep Learning Models for Face Recognition against Image Degradations. arXiv 2017, arXiv:1710.01494v1.
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks; Springer International Publishing: Cham, Switzerland, 2014.
- Guthier, B. Convolutional Neural Networks.
- Sawant, A.; Bhandari, M.; Yadav, R.; Yele, R.; Bendale, M.S. Brain Cancer Detection From MRI: A Machine Learning Approach tensorflow. Int. Res. J. Eng. Technol. 2018, 5, 4.
- Ucar, M.; Ucar, E. Computer-Aided Detection of Lung Nodules in Chest X-rays using Deep Convolutional Neural Networks. Sak. Univ. J. Comput. Inf. Sci 2019, 1–8.
- Sun, J.; Sun, T.; Yuan, Y.; Zhang, X.; Shi, Y.; Lin, Y. Automatic Diagnosis of Thyroid Ultrasound Image Based on FCN-AlexNet and Transfer Learning. In Proceedings of the 2018 IEEE 23rd International Conference on Digital Signal Processing (DSP), Shanghai, China, 19–21 November 2018; pp. 1–5.
- Hosny, K.M.; Kassem, M.A.; Foaud, M.M. Classification of skin lesions using transfer learning and augmentation with Alex-net. PLoS ONE 2019, 14, e0217293.
- Shahzadi, I.; Tang, T.B.; Meriadeau, F.; Quyyum, A. CNN-LSTM: Cascaded Framework For Brain Tumour Classification. In Proceedings of the 2018 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), Sarawak, Malaysia, 3–6 December 2018; pp. 633–637.
- Song, Y.; Zhang, Y.D.; Yan, X.; Liu, H.; Zhou, M.; Hu, B.; Yang, G. Computer-aided diagnosis of prostate cancer using a deep convolutional neural network from multiparametric MRI. J. Magn. Reson. Imaging 2018, 48, 1570–1577.
- Chi, J.; Walia, E.; Babyn, P.; Wang, J.; Groot, G.; Eramian, M. Thyroid Nodule Classification in Ultrasound Images by Fine-Tuning Deep Convolutional Neural Network. J. Digit. Imaging 2017, 30, 477–486.
- Ghosal, P.; Nandanwar, L.; Kanchan, S.; Bhadra, A.; Chakraborty, J.; Nandi, D. Brain Tumor Classification Using ResNet-101 Based Squeeze and Excitation Deep Neural Network. In Proceedings of the 2019 Second International Conference on Advanced Computational and Communication Paradigms (ICACCP), Sikkim, India, 25–28 February 2019; pp. 1–6.
- Chen, X.; Chen, Y.; Ma, C.; Liu, X.; Tang, X. Classification of Pancreatic Tumors Based on MRI Images Using 3D Convolutional Neural Networks. In Proceedings of the 2Nd International Symposium on Image Computing and Digital Medicine, Chengdu, China, 13–14 October 2018.
- Vestias, M.P. A Survey of Convolutional Neural Networks on Edge with Reconfigurable Computing. Algorithms 2019, 12, 154.
- Zhao, X.; Wu, Y.; Song, G.; Li, Z.; Zhang, Y.; Fan, Y. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med. Image Anal. 2018, 43, 98–111.
- Setio, A.A.A.; Ciompi, F.; Litjens, G.; Gerke, P.; Jacobs, C.; van Riel, S.J.; Wille, M.M.W.; Naqibullah, M.; Sánchez, C.I.; van Ginneken, B. Pulmonary Nodule Detection in CT Images: False Positive Reduction Using Multi-View Convolutional Networks. IEEE Trans. Med. Imaging 2016, 35, 1160–1169.
- Kekelidze, M.; D’Errico, L.; Pansini, M.; Tyndall, A.; Hohmann, J. Colorectal cancer: Current imaging methods and future perspectives for the diagnosis, staging and therapeutic response evaluation. World J. Gastroenterol. 2013, 19, 8502–8514.
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. Breast cancer histopathological image classification using Convolutional Neural Networks. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 25 July 2016; pp. 2560–2567.
- Tang, S.; Aoyagi, S.; Ho, Y.; Sato-Shimokawara, E.; Yamaguchi, T. Wearable Sensor Data Visualization based on CNN towards Healthcare Promotion. In Proceedings of the 2020 International Symposium on Community-centric Systems (CcS), Tokyo, Japan, 23–26 September 2020; pp. 1–5.
- Zhao, K.; Jiang, H.; Yuan, T.; Zhang, C.; Jia, W.; Wang, Z. A CNN Based Human Bowel Sound Segment Recognition Algorithm with Reduced Computation Complexity for Wearable Healthcare System. In Proceedings of the 2020 IEEE International Symposium on Circuits and Systems (ISCAS), Seville, Spain, 12–14 October 2020; pp. 1–5.
- Ke, H.; Chen, D.; Shah, T.; Liu, X.; Zhang, X.; Zhang, L.; Li, X. Cloud-aided online EEG classification system for brain healthcare: A case study of depression evaluation with a lightweight CNN. Softw. Pract. Exp. 2020, 50, 596–610.
- Farahani, B.; Barzegari, M.; Shams Aliee, F.; Shaik, K.A. Towards collaborative intelligent IoT eHealth: From device to fog, and cloud. Microprocess. Microsyst. 2020, 72, 102938.
- Hou, J.; Su, H.; Yan, B.; Zheng, H.; Sun, Z.; Cai, X. Classification of tongue color based on CNN. In Proceedings of the 2017 IEEE 2nd International Conference on Big Data Analysis (ICBDA), Beijing, China, 10–12 March 2017; pp. 725–729.
- Liu, C.; Cao, Y.; Alcantara, M.; Liu, B.; Brunette, M.; Peinado, J.; Curioso, W. TX-CNN: Detecting tuberculosis in chest X-ray images using convolutional neural network. In Proceedings of the IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; pp. 2314–2318.
More
Information
Subjects:
Others
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Entry Collection:
Neurodegeneration
Revisions:
2 times
(View History)
Update Date:
16 Mar 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





