Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mi ZHANG | + 1816 word(s) | 1816 | 2022-02-21 10:01:46 | | | |
| 2 | Catherine Yang | -1 word(s) | 1815 | 2022-03-01 03:11:37 | | | | |
| 3 | Catherine Yang | -1 word(s) | 1815 | 2022-03-01 03:12:19 | | | | |
| 4 | Catherine Yang | Meta information modification | 1815 | 2022-03-03 04:31:25 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Zhang, M. A Fiber-Specific Promoter in Cotton. Encyclopedia. Available online: https://encyclopedia.pub/entry/20018 (accessed on 07 February 2026).
Zhang M. A Fiber-Specific Promoter in Cotton. Encyclopedia. Available at: https://encyclopedia.pub/entry/20018. Accessed February 07, 2026.
Zhang, Mi. "A Fiber-Specific Promoter in Cotton" Encyclopedia, https://encyclopedia.pub/entry/20018 (accessed February 07, 2026).
Zhang, M. (2022, March 01). A Fiber-Specific Promoter in Cotton. In Encyclopedia. https://encyclopedia.pub/entry/20018
Zhang, Mi. "A Fiber-Specific Promoter in Cotton." Encyclopedia. Web. 01 March, 2022.
Copy Citation
Cotton fibers, single seed trichomes derived from ovule epidermal cells, are the major source of global textile fibers. Fiber-specific promoters are desirable to study gene function and to modify fiber properties during fiber development. A regulatory sequence from Rho-related GTPase6 (GhROP6) that is predominantly active in initiating and elongating cotton fibers was introduced.
fiber-specific promoter
cotton fiber
1. Introduction
Cotton is a commercial crop providing the most extensively natural fibers used in global textile industries. Currently, genetic engineering has become a powerful approach to benefit cotton production, where transgenic cotton crops account for over 70% [1]. For targeted improvement of fiber properties, it is still a great challenge to allow desirable gene expression in specific tissues and time windows because some modifications, which promote fiber development, would not be favorable to the other tissues or plant growth. For example, improper manipulation of auxin biosynthetic gene iaaM results in abnormalities of the transgenic cottons, although auxin is known to be a vital plant hormone to increase fiber production [2]. In addition, a precise regulation of gene expression in fibers can also facilitate the study of gene expression and function without any systematic influences. Unlike generalized transformation of plants for insect or disease resistance, this must be tissue specific to the surface of developing cotton seeds and time specific to cellular stages. Thus, it is of great importance to isolate and characterize fiber-specific promoters in cotton.
Cotton fibers are differentiated ovule epidermal cells, and their development consists of five continuous stages: initiation, elongation, transition, secondary cell wall synthesis and maturation [3]. Thus far, a series of fiber-related promoters have been characterized with various activities. A part of them is the promoters with activity in fibers from the elongation to the secondary cell wall synthesis stages. E6 and FbL2A are two promoters isolated due to the abundant transcription in cotton fibers [4][5]. E6 and FbL2A show satisfactory specificity in fibers where their activity strengths are roughly equal to one-ninth and one-third, respectively, of that of the constitutive promoter CaMV35S [5]. Another promoter of lipase/hydrolase gene (GhGDSL) also displays fiber-specific activity mainly in the secondary cell wall synthesis stage [6]. Similar activity behavior is observed in promoters of chitinase-like protein (GhCTL) and TCP transcriptional factor (GbTCP), except for some additional activity in anthers, xylems or young cotyledons and roots [7][8]. In addition, the regulatory sequences of RAC13, CelA1 and LTP also have activity in fibers of the elongation and secondary cell wall synthesis stages [9].
2. GhROP6 Is Preferentially Expressed in Fiber Cells
The expression pattern of GhROP6 in cotton tissues was first examined according to the previous RNA-seq data [10]. Two copies (Gh_A01G1392 and Gh_D01G1636) of GhROP6 showed a preferential expression in developing ovules and fibers apart from the highest transcription in petals (Figure 1A). Moreover, both, except for GhROP6-A at 10 DPA, showed a considerably higher expression in fiber tissues than in ovule tissues of 5 to 25 DPA (Figure 1A). The expression profile of GhROP6 was further confirmed by quantitative RT-PCR (Figure 1B). Similarly, GhROP6 was highly expressed in separated fibers and petals, although the transcript was detected in all tested tissues. The expression in 9-DPA fibers was the highest one, versus that in petals and in 15-DPA fibers. In addition, the expression in fibers was roughly three-fold higher than the separated ovule at 9 and 15 DPA. These data suggest that GhROP6 is preferentially expressed in fibers of the early developmental stage.
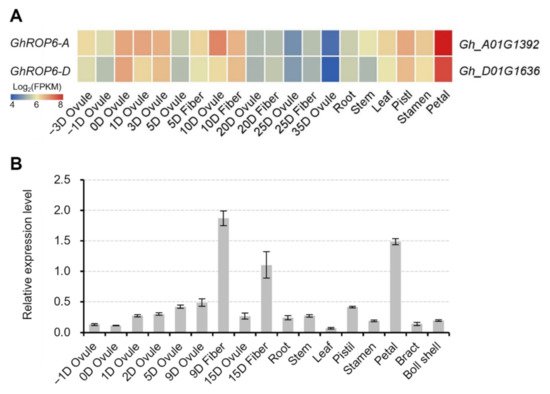
Figure 1. Expression profile of GhROP6 in cotton tissues. (A) Heatmap of expression pattern of GhROP6 copies. GhROP6-A (Gh_A01G1392) and GhROP6-D (Gh_D01G1636) are two copies of GhROP6 in A- and D-subgenome of upland cotton. The heatmap was generated from the released data [10]. (B) Transcription level of GhROP6 (in arbitrary units) normalized to that of GhHIS3. Error bars represent standard deviation of three technical repeats. Samples at −3 to 3 DPA (A) or −1 to 5 DPA (B) were fiber-bearing ovules.
3. Molecular Characterization of proGhROP6::GUS Transgenic Cotton
15 transgenic cotton plants were obtained and confirmed by detection of the NPTII (kanamycin resistance) gene (Figure 2A) in the genome. GUS activity was then examined in 0-DPA ovules of these transgenic cottons. Eight plants showed visible staining in the ovule surface, and five (#1, #5, #19, #21 and #100) of them were further selected to analyze GUS activity in the other tissues (Figure 2B). Except for transformant #100 that showed strong staining in all tested tissues, all transformants showed no discernable activity in leaves (even in leaf trichomes), stems, petal, stamens, pistils and roots (Figure 2B). The absent activity in petals could be attributed to exclusion of the corresponding tissue-specific elements in the GhROP6 promoter. These results indicate that the proGhROP6 has strict activity in developing ovules.
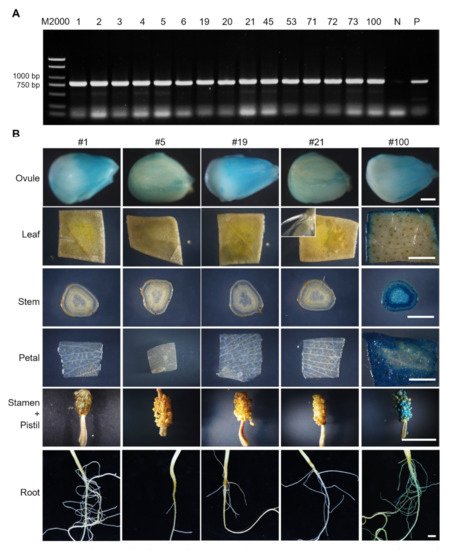
Figure 2. Identification of proGhROP6::GUS transgenic cotton. (A) PCR identification of introduction of selection gene NPTII. Number of transformants is shown in each lane. N represents the negative control. P represents the positive control of a NPTII-containing plasmid. (B) GUS staining in ovules, leaves, stems, petals, stamens, pistils, and roots. Floral organs were harvested at 0 DPA. The inset shows leaf trichomes. Scale bars represent 500 µm (ovule images) or 5 mm (the others).
4. The proGhROP6 Drives Normal Transcription of GUS Reporter Gene
The inclusion of the first exon and the partial first intron of GhROP6 in the proGhROP6 sequence raised a concern about aberrant mRNA splicing of the target gene placed downstream. We thus analyzed three possible transcripts, starting from the predicted 5’-UTR, the first exon of GhROP6, and the start codon of GUS with selective primer pairs (Figure 3A) in tissues of proGhROP6::GUS transgenic cotton. GUS transcript was detected in cDNA extracted from 2-DPA ovules, while two bands were amplified out with the upper stream primer locating at the first exon of GhROP6 (Figure 3B). We sequenced them, and the results showed that the long one was the predicted transcript and the short one had a splicing in the partial intron region of the proGhROP6 promoter. No other splicing was found in the coding region of GUS. We did not detect any specific amplicon with the primer located at the 5’-UTR (Figure 3B). No other specific amplicons, except for faint transcription of GUS in the stem, were detected with all three primer pairs in these tissues (Figure 3B–E). These results suggest that the detected GUS activity on ovule surface is due to the correct transcription of GUS gene, and the proGhROP6 does not direct any abnormal processing of GUS in cotton tissues.
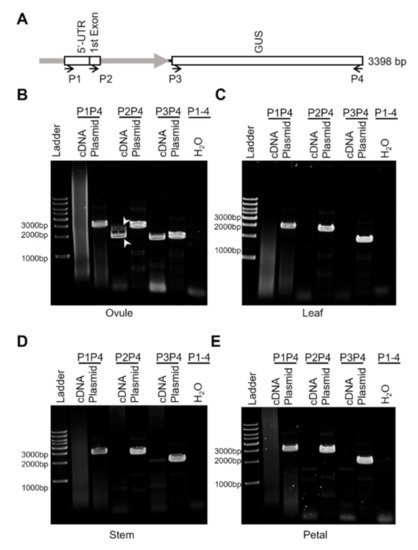
Figure 3. Transcription analysis of proGhROP6::GUS in the transgenic cotton. (A) Diagram of proGhROP6::GUS and primer location. proGhROP6 contains the predicted 5′-UTR, the first exon, and part of the first intron of GhROP6. P1 to P4, primers for RT-PCR analysis; Plasmid, positive control using the plasmid as the template; H2O, negative control using the distilled water as the template. (B–E) RT-PCR analysis of proGhROP6::GUS transcripts. cDNA was synthesized using RNA extracted from 2-DPA ovules (B), leaves (C), stems (D) and petals (E). Arrowheads indicate the two transcripts of proGhROP6::GUS during RNA processing.
5. Activity of the proGhROP6 in Developing Ovules
All three transformants showed a similar GUS expression pattern (Figure 4). GUS staining on the ovule surface was of peak value at 0 or 2 DPA and then decreased after 5 DPA. This tendency was obvious in transformant #19 and #21, where the staining was weak at 10 to 20 DPA. The GUS activity was mainly enriched in ovule epidermal cells before anthesis. However, dotted blue staining on the ovule surface (insets in Figure 4) indicated stronger activity in fiber cells after anthesis, which was further confirmed in ovule sections. Thus, the promoter of GhROP6 is mainly active in fiber cells of the initiation stage.
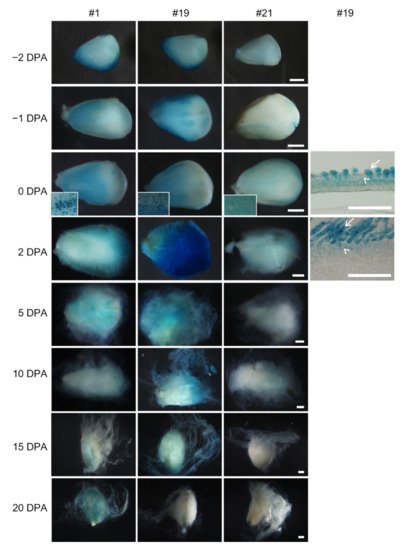
Figure 4. GUS activity in developing ovules of proGhRop6::GUS transgenic cotton. Gus staining in ovules of transformant #19 were further observed through paraffin-embedded sections. Arrows indicate fiber cells, and arrowheads indicate non-fiber cells in ovule epidermis. Scale bars represent 500 µm (ovule images) or 50 µm (sections).
6. The proGhROP6 Is a Weak Promoter Active in Fiber Initiation Stage
In order to verify strength of the proGhROP6 activity, we quantified GUS activity in developing ovules (−2 to 2 DPA) and fibers (after 2 DPA) of proGhROP6::GUS transformant #1 (Figure 5A). The GUS activity reached the highest level at 0 DPA and then decreased to a base level at 10 DPA, in which the value was similar to that at −2 and 20 DPA. Approximately, the 0 DPA value was seven times as high as the base level. GUS activity of proGhROP6::GUS ovules was nearly one-twenty-fifth of that in 35S::GUS ovules (Figure 5B). These data suggest that proGhROP6 is a mild promoter active in the fiber initiation stage.
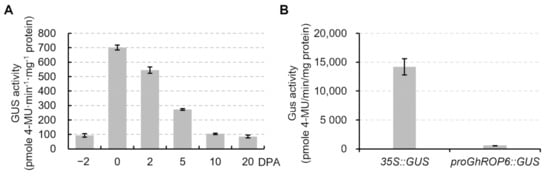
Figure 5. Strength of GUS activity in proGhROP6::GUS ovules. (A) GUS activity in developing ovules. Samples from −2 to 2 DPA were fiber-bearing ovules, and those from 5 to 20 DPA were separated fibers. (B) Comparison of GUS activity in 0 DPA ovules between proGhROP6::GUS and 35S::GUS cottons. Error bars represent standard deviations of three biological replicates.
7. Responses of the GhROP6 Promoter to Plant Hormones
A series of cis-acting regulatory elements in response to plant hormones auxin, gibberellin, jasmonate, and salicylic acid were detected (Figure 6A) using the database PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 26 October 2021)) or the reported motifs [11][12]. This implied that this proGhROP6 might be responsive to these hormones. To test it, wild-type ovules were treated with these plant hormones to analyze expression of GhROP6. GhROP6 transcription was slightly decreased in the presence of IAA at three concentrations (0.5, 5.0 and 50 μM), but the responses were not dose-dependent (Figure 6B). No discernable change was detected as compared to wild-type ovules (Figure 6C), suggesting that the induced decrease in GhROP6 expression by auxin is limited. Decreased expression of GhROP6 was also detected in gibberellin acid (GA3)-treated ovules, but only at the high concentration of 50 μM (Figure 6D). Similar results were also detected in ovules treated with salicylic acid (Figure 6F). In contrast, no trend of GhROP6 expression was seen in methyl jasmonate (MeJA)-treated ovules (Figure 6E). While at the highest concentration, a large portion (from 79% to 91%) of ovules with slightly decreased GUS staining were observed in the presence of IAA, GA3 or SA, and the GUS signal in MeJA treatment was undistinguishable compared with the control (Figure 6G–K). All these treatments did not change the enriched activity of GhROP6 promoter in fiber cells (insets in Figure 6G–K).
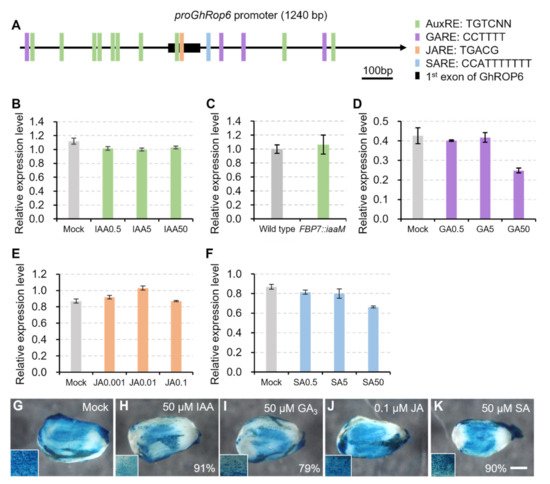
Figure 6. Activity of GhROP6 promoter in response to different plant hormones. (A) Response elements for different plant hormones in the GhROP6 promoter region. (B) Transcription level of GhROP6 in wild-type ovules treated with different concentration of IAA. (C) Transcription level of GhROP6 in FBP7::iaaM and wild-type ovules. (D) Transcription level of GhROP6 in wild-type ovules treated with different concentration of GA3. (E) Transcription level of GhROP6 in wild-type ovules treated with different concentration of MeJA. (F) Transcription level of GhROP6 in wild-type ovules treated with different concentration of SA. Transcription level (in arbitrary units) was normalized to that of GhHIS3. Error bars represent standard deviation of three repeats. (G–K) GUS staining in proGhROP6::GUS transgenic ovules after treatment with different plant hormones. Over twenty ovules of transformant #1 were harvested for each treatment. Percentage of ovules with reduced GUS staining are shown. GUS staining of ovules in the lower concentrations were indistinguishable from the control. Insets show enlargement of ovule surface. The scale bars represent 500 μm. Ovules at 0 DPA, except for those in (C), were treated with different plant hormones for 6 h.
References
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537.
- Zhang, M.; Zheng, X.; Song, S.; Zeng, Q.; Hou, L.; Li, D.; Zhao, J.; Wei, Y.; Li, X.; Luo, M.; et al. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat. Biotechnol. 2011, 29, 453–458.
- Haigler, C.H.; Betancur, L.; Stiff, M.R.; Tuttle, J.R. Cotton fiber: A powerful single-cell model for cell wall and cellulose research. Front. Plant Sci. 2012, 3, 104.
- John, M.E.; Crow, L.J. Gene expression in cotton (Gossypium hirsutum L.) fiber: Cloning of the mRNAs. Proc. Natl. Acad. Sci. USA 1992, 89, 5769–5773.
- Rinehart, J.A.; Petersen, M.W.; John, M.E. Tissue-specific and developmental regulation of cotton gene FbL2A. Demonstration of promoter activity in transgenic plants. Plant Physiol. 1996, 112, 1331–1341.
- Yadav, V.K.; Yadav, V.K.; Pant, P.; Singh, S.P.; Maurya, R.; Sable, A.; Sawant, S.V. GhMYB1 regulates SCW stage-specific expression of the GhGDSL promoter in the fibres of Gossypium hirsutum L. Plant Biotechnol. J. 2017, 15, 1163–1174.
- Zhang, D.; Hrmova, M.; Wan, C.-H.; Wu, C.; Balzen, J.; Cai, W.; Wang, J.; Densmore, L.D.; Fincher, G.B.; Zhang, H.; et al. Members of a new group of chitinase-like genes are expressed preferentially in cotton cells with secondary walls. Plant Mol. Biol. 2004, 54, 353–372.
- Hao, J.; Tu, L.; Hu, H.; Tan, J.; Deng, F.; Tang, W.; Nie, Y.; Zhang, X. GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 2012, 63, 6267–6281.
- Chen, J.; Burke, J.J. Developing fiber specific promoter-reporter transgenic lines to study the effect of abiotic stresses on fiber development in cotton. PLoS ONE 2015, 10, e0129870.
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotech. 2015, 33, 524–530.
- Mena, M.; Cejudo, F.J.; Isabel-Lamoneda, I.; Carbonero, P. A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol. 2002, 130, 111–119.
- Sghaier, N.; Ben Ayed, R.; Gorai, M.; Rebai, A. Prediction of auxin response elements based on data fusion in Arabidopsis thaliana. Mol. Biol. Rep. 2018, 45, 763–772.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
923
Revisions:
4 times
(View History)
Update Date:
03 Mar 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




