Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Matthew Winans | + 1783 word(s) | 1783 | 2022-02-22 04:37:50 | | | |
| 2 | Rita Xu | Meta information modification | 1783 | 2022-02-22 05:18:56 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Winans, M. Yeast Hybrids in Brewing. Encyclopedia. Available online: https://encyclopedia.pub/entry/19724 (accessed on 07 February 2026).
Winans M. Yeast Hybrids in Brewing. Encyclopedia. Available at: https://encyclopedia.pub/entry/19724. Accessed February 07, 2026.
Winans, Matthew. "Yeast Hybrids in Brewing" Encyclopedia, https://encyclopedia.pub/entry/19724 (accessed February 07, 2026).
Winans, M. (2022, February 22). Yeast Hybrids in Brewing. In Encyclopedia. https://encyclopedia.pub/entry/19724
Winans, Matthew. "Yeast Hybrids in Brewing." Encyclopedia. Web. 22 February, 2022.
Copy Citation
Microbiology has long been a keystone in fermentation, and innovative yeast molecular biotechnology continues to represent a fruitful frontier in brewing science. Consequently, modern understanding of brewer’s yeast has undergone significant refinement over the last few decades.
hybrid
lager
yeast
introgression
interspecific
domestication
1. Species of Saccharomyces
Saccharomyces cerevisiae may be one of the oldest domesticated organisms known to humans. Domestication events imposed on brewing strains of the budding yeast species S. cerevisiae resulted in unique strains similar to the divergence seen in animal lineages of Canis familiaris breeds or the plant lineage of Brassica oleracea foods. It has been suggested that S. cerevisiae behaved as a synanthropic species, following human settlements as a commensal organism residing in gardens and vineyards, although the time period and location of the yeast’s origins has been the subject of much debate throughout history. Domesticated Saccharomyces brewing strains feature flocculation capabilities, fast fermentation rates, malt sugar utilization, pleasant aromas, and are largely negative for production of phenolic off flavors (POF) [1][2].
Nearly two centuries have passed since the first accessible description was produced regarding brewer’s yeast and its recognition in fermentation [3][4]. Recently, phylogenic research utilizing genomics and modern molecular biology techniques has shed some light on the historically convoluted nomenclature surrounding this budding yeast. Genomic analysis of the Saccharomyces genus has consolidated many variations into eight individual species: S. cerevisiae, S. paradoxus (syn. S. cariocanus, S. cerevisiae var. tetraspora, S. cerevisiae var. terrestris, S. douglasii), S. uvarum (syn. S. bayanus var. uvarum), S. mikatae, S. kudriavzevii, S. arboricola (syn. S. arboricolus), S. eubayanus, and S. jurei [3][5][6][7][8][9][10][11][12][13][14][15][16][17][18][19] (Table 1). Moreover, two natural hybrids are recognized in the Saccharomyces clade: S. pastorianus (syn. S. carlsbergensis, S. monacensis) and S. bayanus [20][21][22]. Most modern lager fermentations utilize S. patorianus yeasts.
Table 1. Current Saccharomyces Yeast Species History.
| Saccharomyces | Described | Substrate | Location | Reference |
|---|---|---|---|---|
| cerevisiae | 1838 | Beer | Germany | [4] |
| uvarum | 1898 | Ribes rubrum, redcurrant juice | South Holland, The Netherlands | [16][17] |
| paradoxus | 1914 | Tree sap | Russia | [15] |
| kudriavzevii | 1991 | Decayed leaf | Japan | [18] |
| mikatae | 1993 | Decayed leaf | Japan | [19] |
| arboricola | 2008 | Fagaceae spp. | West China | [6] |
| eubayanus | 2011 | Nothofagus spp. & parasitic fungi Cyttaria spp. | Andean, Patagonia | [11] |
| jurei | 2017 | Quercus robur | Saint Auban, France | [14] |
2. Hybrid Nature of Yeast
Interspecific hybrids are not unique to lager brewing. For example, the livestock and agricultural industries commonly employ selective breeding to alter species’ properties or increase yields [23][24][25]. A time-honored showpiece of hybrid vigor is the mule, a great pack animal known for its hardiness and longevity. For over 4000 years, the mule has been bred as the hybrid progeny of a male donkey and a female horse. Since the early 1900s, maize has been hybridized to increase yields and introduce biodiversity [26]. Similarly, hybrid yeasts have been isolated from fermentation processes on numerous occasions [27] (Figure 1). A hybrid between S. cerevisiae and S. kudriavzevii was isolated from Belgian Trappist beers [28]. Popular in wine production, strain VIN7 is a hybrid of S. cerevisiae and S. kudriavzevii [29]. Other interspecific S. cerevisiae and S. uvarum hybrids are also regularly used for production of wines [29][30]. Spontaneous fermentations have yielded Pichia apotheca, a hybrid of P. membranifaciens and an unknown species [31].
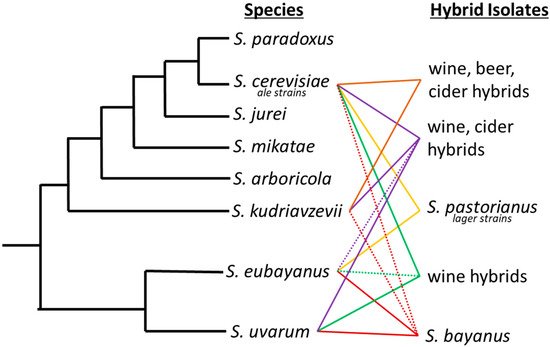
Figure 1. Saccharomyces phylogenetic tree with industrially important hybrids. Industrial hybrids are listed to the right by the fermentation they have predominately been associated with. Solid lines between two species signifies the interspecific hybrids and dashed lines denote introgression from a third or fourth species that may not always be present in each hybrid strain.
The mule of the brewing industry is the lager yeast S. pastorianus, an interspecific hybrid that produces the lion’s share, in volume, of the global beer production. Although its use is widespread, the biodiversity is limited to two main lineages, Saaz/group I (syn. S. carlsbergensis, (L12: Noble, Imperial Yeast Culture Collection, type strain CBS1513) and Frohberg/group II (L13: Global, Imperial Yeast Culture Collection, type strain-Weihenstephan 34/70). Saaz and Frohberg lineages vary in their genomic composition from each parent species, S. eubayanus and S. cerevisiae, which influences important fermentation characteristics. Genomic analysis demonstrated a genomic composition of 1:2 S. cerevisiae to S. eubayanus sub genome in the Saaz lineage and 2:2 S. cerevisiae to S. eubayanus sub genome in the Frohberg lineage supporting the traditional designations used by brewers [32][33][34].
3. Novel Hybrid Development
The first yeast breeding experiments aimed at combining desirable traits of brewing strains were conducted by Ojvind Winge during his tenure at the Carlsberg Laboratory in the 1930s [35]. Hybrid yeast development has been carried out for over half a century since then, aimed predominantly at increasing attenuation and fermentation rates via intraspecific crosses with ale and lab strains [36][37][38]. Modern fermentations benefit from many innate and acquired hybrids that have been isolated or developed [11][27][28][39][40][41][42][43][44][45][46][47][48][49][50]. Early efforts in brewing science established the fundamentals necessary to explore the phylogeny, genomics, and strain development for Saccharomyces fermentation. During typical rich nutrient propagations of yeast in a brewing environment, mother cells reproduce asexually to bud off small daughter clones (Figure 2). Under poor nitrogen conditions, such as proline, yeast growth changes to a pseudohyphal form [51][52]. The complete absence of a nitrogen source and the presence of a non-fermentable carbon source, such as acetate, will sporulate yeast cells [53]. Sporulation transforms the cell wall into the ascus, or sack, that holds four spores termed a tetrad. Analogous to the human egg and sperm, these spores divide equally into mating types as either a or α [54]. When conditions improve for yeast growth, new haploid (1n) yeast can conjugate with the opposite mating type yeast as they form a shmoo.
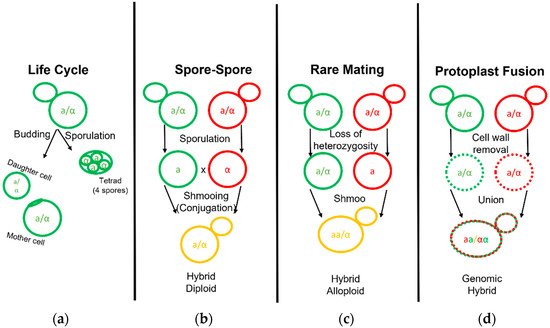
Figure 2. Life and Mating of Saccharomyces Yeast. Diagram pertaining to the clonal growth typical of yeast fermentation cultures and the various known techniques employed to generate yeast hybrids. (a) Diploid yeast cells may bud and grow clonally to form a mother and daughter cell or undergo sporulation to form a tetrad. (b) Yeast hybridization may form by direct spore to spore mating. (c) Yeast hybridization may form by rare mating events in which one or both diploid parent cells gain competency by becoming hemizygous or MATa/MATa and MATα/MATα diploids. (d) Yeast hybridization may also form by fusion of two separate yeast cell protoplasts with their cell wall removed.
Interspecific hybridization is seen as a valuable tool for yeast strain development, enabling the combination and enhancement of characteristics from both parental strains or species [55]. The development of hybrids is executed via three primary methodologies: spore–spore mating, rare mating, and protoplast fusion (Figure 2). Spore to spore mating is most similar to what would be considered natural mating, as outlined in Figure 2b. This approach bears a high success rate, high genomic stability, and can avoid the aid of selection markers such as drug resistance or autotrophies. Rare mating utilizes a described spontaneous loss of heterozygosity at the mating type locus. Normal diploid cells carry two sets of chromosomes with both the MATa and MATα genetic alleles and do not respond to sex pheromones for mating purposes. The spontaneous loss of either sex allele tolerates yeast mating to a yeast cell of complimentary sex. This results in yeast with high chromosome counts, influencing gene dosage during cellular processes and partially explains the outperformance over a diploid yeast of the same background [55]. Rare mating, as the name implies, is uncommon and selection markers are needed to perform this technique. The frequency of rare mating is estimated to occur in 1 out of 10 million cells [56]. This procedure is beneficial in overcoming poor sporulation but produces hybrids prone to high genomic instability. Lastly, protoplast fusion is performed by removing the cell wall and fusing the protoplasts of two cells together before the cell wall is repaired. This technique generates cells with a high chromosome copy number and higher genomic instability but overcomes low sporulation. This technique can be used in the laboratory to combine yeast from different genera such as the brewer’s yeast S. cerevisiae and other yeast outside of the Saccharomyces genera which are otherwise incompatible [57]. Protoplast fusion is considered genetic engineering in many parts of the world. Recent select investigations into Saccharomyces hybrid application in beverage fermentations are listed in table format (Table 2).
Utilizing advanced molecular biology techniques for the modification of yeast strains presents an ongoing endeavor by many academic labs and some commercial yeast laboratories. Several research groups have developed protocols that use plasmids carrying genetic markers for drug resistance or functional enzymes that target the mating system in yeast. Industrial strains are well-known for their poor ability to adhere to laboratory techniques including sporulation and transformation [58]. One strategy developed at the University of Wisconsin generates allotetraploid strains of prototrophic yeast without the need for sporulation or modification to the nuclear genome of parental yeast strains [59]. This method leverages a series of inducible plasmids coined HyPr (Hybrid Production) containing compilatory drug markers and an inducible HO cassette. HO encodes an endonuclease that performs a double stranded break in the DNA that determines the Saccharomyces sex type. To repair this damage, the yeast cell will copy the silenced sex type already present in the genome, effectively performing a sex change and allowing the cell to mate with other cells of the opposite mating type. Using drug resistance markers and the HO cassette, hybrids are produced and serial growth in the absence of drug selection purges cells of exogenous DNA [59].
Currently, the options for strain selection in S. cerevisiae yeast are plentiful, but the criteria for fermentation performance in the brewing environment remains selective. Strains are employed largely by beer style, equipment availability, and supporting knowledge base. Certain beer styles also contain defining features from specific yeast flavor active molecules [60]. Banana and clove flavors are derived from isoamyl acetate and 4-vinyl guaiacol (4VG) in Weissbier [60][61][62]. Hybridization of yeast bears several advantages in brewing to include transgressive phenotypes such as increased ethanolic fermentation performance or stress tolerance, shifting fermentation temperatures beyond traditional inhibitory conditions, and creating a mosaic blend of parental fermentation profiles [24][49][55][63][64]. Targets of yeast hybridization may include increased formation of glycerol for an enhanced mouthfeel, alternative carbon source metabolic ability, reduced off-flavor production, increased formation of antioxidants that increase beer flavor stability, or increase production of yeast longevity molecules such as trehalose. The methodologies to create yeast hybrids vary in their specificity to target genetic or phenotypic results, but efforts to harness yeast hybrids in brewing broadly increase the biodiversity of fermentation yeast, add depth to the complexity of fermentation profiles, and advance brewing science knowledge.
Table 2. Interspecific Yeast Hybrids in Fermentation.
| History | Parents | Reference |
|---|---|---|
| Isolated | S. cerevisiae × S. eubayanus | [11] |
| Isolated | S. cerevisiae ×S. eubayanus × S. uvarum | [39] |
| Isolated | S. cerevisiae × S. uvarum | [65] |
| Isolated | S. cerevisiae × S. kudriavzevii | [27][28][40] |
| Isolated | S. uvarum × S. eubayanus | [27][40] |
| Developed | S. cerevisiae × S. eubayanus | [41][43][44][45][66] |
| Developed | S. cerevisiae × S. mikatae | [46] |
| Developed | S. cerevisiae ×S. kudriavzevii × S. paradoxus | [47] |
| Developed | S. cerevisiae × S. kudriavzevii | [50] |
| Developed | S. cerevisiae × S. arboricola | [46][49] |
| Developed | S. cerevisiae × S. jurei | [67] |
References
- Gallone, B.; Steensels, J.; Baele, G.; Maere, S.; Verstrepen, K.J.; Prahl, T.; Soriaga, L.; Saels, V.; Herrera-Malaver, B.; Merlevede, A.; et al. Domestication and Divergence of Saccharomyces cerevisiae Beer Yeasts. Cell 2016, 166, 1397–1410.e16.
- Preiss, R.; Tyrawa, C.; Krogerus, K.; Garshol, L.M.; Van Der Merwe, G. Traditional Norwegian Kveik are a genetically distinct group of domesticated Saccharomyces cerevisiae brewing yeasts. Front. Microbiol. 2018, 9, 2137.
- Pasteur, L. Nouveaux faits concernant l’histoire de la fermentation alcoolique. Comptes Rendus Chim. 1858, 47, 1011–1013.
- Meyen, F.J.F. Jahresbericht über die Resultate der Arbeiten im Felde der physiologischen Botanik von dem Jahre 1837. In Berlin: Nicolai’sche Buchhandlung; Nicolai: Berlin, Germany, 1838; pp. 1–186.
- Martini, A.V.; Martini, A. Three newly delimited species of Saccharomyces sensu stricto. Antonie Van Leeuwenhoek 1987, 53, 77–84.
- Wang, S.A.; Bai, F.Y. Saccharomyces arboricolus sp. nov., a yeast species from tree bark. Int. J. Syst. Evol. Microbiol. 2008, 58, 510–514.
- Naumova, E.S.; Roberts, I.N.; James, S.A.; Naumov, G.I.; Louis, E.J. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int. J. Syst. Evol. Microbiol. 2015, 50, 1931–1942.
- Naumov, G.I.; Naumova, E.S.; Hagler, A.N.; Mendonça-Hagler, L.C.; Louis, E.J. A new genetically isolated population of the Saccharomyces sensu stricto complex from Brazil. Antonie Van Leeuwenhoek 1995, 67, 351–355.
- Naumov, G.I.; Lee, C.F.; Naumova, E.S. Molecular genetic diversity of the Saccharomyces yeasts in Taiwan: Saccharomyces arboricola, Saccharomyces cerevisiae and Saccharomyces kudriavzevii. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2013, 103, 217–228.
- Naumov, G.I.; Naumova, E.S.; Louis, E.J. Two New Genetically Isolated Popoulations of the Saccharomyces Sensu Stricto Complex from Japan. J. Gen. Appl. Microbiol. 1995, 41, 499–505.
- Libkind, D.; Hittinger, C.T.; Valério, E.; Gonçalves, C.; Dover, J.; Johnston, M.; Gonçalves, P.; Sampaio, J.P. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 14539–14544.
- Boynton, P.J.; Greig, D. The ecology and evolution of non-domesticated Saccharomyces species Primrose. Yeast 2014, 31, 449–462.
- Sipiczki, M. Interspecies hybridisation and genome chimerisation in Saccharomyces: Combining of gene pools of species and its biotechnological perspectives. Front. Microbiol. 2018, 9, 3071.
- Naseeb, S.; James, S.A.; Alsammar, H.; Michaels, C.J.; Gini, B.; Nueno-Palop, C.; Bond, C.J.; McGhie, H.; Roberts, I.N.; Delneri, D. Saccharomyces jurei sp. Nov., isolation and genetic identification of a novel yeast species from Quercus robur. Int. J. Syst. Evol. Microbiol. 2017, 67, 2046–2052.
- Batschinskaya, A. Entwicklungsgeschichte und Kultur des neuen Hefepilzes Saccharomyces paradoxus. J. Microbiol. Epidemiol. Immunobiol. 1914, 1, 231–247.
- Beijerinck, M. Uber Regeneration der Sporenbildung bei Alkohol Hefen, wo diese Funktion im Verchwinden begriffen ist. Cent. Bakt 1898, 4, 657–731.
- Beijerinck, M. Sur la generation de la faculte de produire des spores chez des levures en voie de la perdre. Extr. Des Arch. Neelandaises Des Sci. Exactes Nat. Ser. II 1898, 2, 1–81.
- Kaneko, Y.; Banno, I. Re-examination of Saccharomyces bayanus strains by DNA-DNA hybridization and electrophoretic karyotyping. IFO Res. Commun. 1991, 15, 30–41.
- Yamada, Y.; Mikata, K.; Banno, I. Reidentification of 121 strains of the genus Saccharomyces. Bull JFCC 1993, 9, 95–119.
- Masneuf, I.; Hansen, J.; Groth, C.; Piskur, J.; Dubourdieu, D. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Environ. Microbiol. 1998, 64, 3887–3892.
- Nguyen, H.V.; Legras, J.L.; Neuvéglise, C.; Gaillardin, C. Deciphering the hybridisation history leading to the lager lineage based on the mosaic genomes of Saccharomyces bayanus strains NBRC1948 and CBS380 T. PLoS One 2011, 6, e25821.
- Querol, A.; Bond, U. The complex and dynamic genomes of industrial yeasts: MINIREVIEW. FEMS Microbiol. Lett. 2009, 293, 1–10.
- Chen, Z.J. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 2013, 14, 471–482.
- Fu, D.; Xiao, M.; Hayward, A.; Jiang, G.; Zhu, L.; Zhou, Q.; Li, J.; Zhang, M. What is crop heterosis: New insights into an old topic. J. Appl. Genet. 2015, 56, 1–13.
- Schnable, P.S.; Springer, N.M. Progress toward understanding heterosis in crop plants. Annu. Rev. Plant Biol. 2013, 64, 71–88.
- Crow, J.F. 90 Years Ago: The Beginning of Hybrid Maize. Genetics 1998, 148, 923–928.
- Gallone, B.; Steensels, J.; Mertens, S.; Dzialo, M.C.; Gordon, J.L.; Wauters, R.; Theßeling, F.A.; Bellinazzo, F.; Saels, V.; Herrera-Malaver, B.; et al. Interspecific hybridization facilitates niche adaptation in beer yeast. Nat. Ecol. Evol. 2019, 3, 1562–1575.
- González, S.S.; Barrio, E.; Querol, A. Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing. Appl. Environ. Microbiol. 2008, 74, 2314–2320.
- Borneman, A.R.; Desany, B.A.; Riches, D.; Affourtit, J.P.; Forgan, A.H.; Pretorius, I.S.; Egholm, M.; Chambers, P.J. The genome sequence of the wine yeast VIN7 reveals an allotriploid hybrid genome with Saccharomyces cerevisiae and Saccharomyces kudriavzevii origins. FEMS Yeast Res. 2012, 12, 88–96.
- Le Jeune, C.; Lollier, M.; Demuyter, C.; Erny, C.; Legras, J.-L.; Aigle, M.; Masneuf Pomarède, I. Characterization of natural hybrids of Saccharomyces cerevisiae and Saccharomyces bayanus var. uvarum. FEMS Yeast Res. 2007, 7, 540–549.
- Heil, C.S.; Burton, J.N.; Liachko, I.; Friedrich, A.; Hanson, N.A.; Morrise, C.L.; Schacherer, J.; Shendurer, J.; Thomas, J.H.; Maitreya, J. Dunham Identification of a novel interspecific hybrid yeast from a metagenomic spontaneously inoculated beer sample using Hi-C. Yeast 2018, 35, 71–84.
- Liti, G.; Peruffo, A.; James, S.A.; Roberts, I.N.; Louis, E.J. Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast 2005, 22, 177–192.
- Dunn, B.; Sherlock, G. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 2008, 18, 1610–1623.
- Glendinning, T. Some Practical Aspects of the Fermentable Matter. J. Fed. Inst. Brew. 1898, 5, 20–34.
- Barnett, J.A. A history of research on yeasts 10: Foundations of yeast genetics. Yeast 2007, 24, 799–845.
- Hammond, B.J.R.M.; Eckersley, K.W. Fermentation Properties of Brewing Yeast with Killer Character. J. Insta. Brew. 1984, 90, 167–177.
- Johnston, J.R. Breeding yeasts for brewing. J. Inst. Brew. 1965, 71, 135–137.
- Spencer, J.F.T.; Spencer, D.M. Hybridization of non-sporulating and weakly sporulating strains of brewer’s and distiller’s yeasts. J. Inst. Brew. 1977, 83, 287–289.
- Peŕez-Través, L.; Lopes, C.A.; Querol, A.; Barrio, E. On the complexity of the Saccharomyces bayanus taxon: Hybridization and potential hybrid speciation. PLoS ONE 2014, 9, e93729.
- Langdon, Q.K.; Peris, D.; Baker, E.C.P.; Opulente, D.A.; Nguyen, H.V.; Bond, U.; Gonçalves, P.; Sampaio, J.P.; Libkind, D.; Hittinger, C.T. Fermentation innovation through complex hybridization of wild and domesticated yeasts. Nat. Ecol. Evol. 2019, 3, 1576–1586.
- Mertens, S.; Steensels, J.; Saels, V.; De Rouck, G.; Aerts, G.; Verstrepen, K.J. A large set of newly created interspecific Saccharomyces hybrids increases aromatic diversity in lager beers. Appl. Environ. Microbiol. 2015, 81, 8202–8214.
- Magalhães, F.; Krogerus, K.; Vidgren, V.; Sandell, M.; Gibson, B. Improved cider fermentation performance and quality with newly generated Saccharomyces cerevisiae × Saccharomyces eubayanus hybrids. J. Ind. Microbiol. Biotechnol. 2017, 44, 1203–1213.
- Krogerus, K.; Magalhães, F.; Vidgren, V.; Gibson, B. New lager yeast strains generated by interspecific hybridization. J. Ind. Microbiol. Biotechnol. 2015, 42, 769–778.
- Krogerus, K.; Arvas, M.; De Chiara, M.; Magalhães, F.; Mattinen, L.; Oja, M.; Vidgren, V.; Yue, J.X.; Liti, G.; Gibson, B. Ploidy influences the functional attributes of de novo lager yeast hybrids. Appl. Microbiol. Biotechnol. 2016, 100, 7203–7222.
- Hebly, M.; Brickwedde, A.; Bolat, I.; Driessen, M.R.M.; de Hulster, E.A.F.; van den Broek, M.; Pronk, J.T.; Geertman, J.M.; Daran, J.M.; Daran-Lapujade, P. S. cerevisiae × S. eubayanus interspecific hybrid, the best of both worlds and beyond. FEMS Yeast Res. 2015, 15, 1–14.
- Nikulin, J.; Krogerus, K.; Gibson, B. Alternative Saccharomyces interspecies hybrid combinations and their potential for low-temperature wort fermentation. Yeast 2018, 35, 113–127.
- Bellon, J.R.; Eglinton, J.M.; Siebert, T.E.; Pollnitz, A.P.; Rose, L.; De Barros Lopes, M.; Chambers, P.J. Newly generated interspecific wine yeast hybrids introduce flavour and aroma diversity to wines. Appl. Microbiol. Biotechnol. 2011, 91, 603–612.
- Bellon, J.R.; Schmid, F.; Capone, D.L.; Dunn, B.L.; Chambers, P.J. Introducing a New Breed of Wine Yeast: Interspecific Hybridisation between a Commercial Saccharomyces cerevisiae Wine Yeast and Saccharomyces mikatae. PLoS ONE 2013, 8, e62053.
- Winans, M.J.; Yamamoto, Y.; Fujimaru, Y.; Kusaba, Y.; Gallagher, J.E.G.; Kitagaki, H. Saccharomyces arboricola and Its Hybrids’ Propensity for Sake Production: Interspecific hybrids reveal increased fermentation abilities and a mosaic metabolic profile. Fermentation 2020, 6, 14.
- Bizaj, E.; Cordente, A.G.; Bellon, J.R.; Raspor, P.; Curtin, C.D.; Pretorius, I.S. A breeding strategy to harness flavor diversity of Saccharomyces interspecific hybrids and minimize hydrogen sulfide production. FEMS Yeast Res. 2012, 12, 456–465.
- Gancedo, J.M. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001, 25, 107–123.
- Jones, M.; Pierce, J. Absorption of Amino Acids from Wort by Yeast. J. Inst. Brew. 1964, 70, 307–315.
- Esposito, R.E.; Klapholz, S. Meiosis and Ascospore Development. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance; Strathern, J.N., Jones, E.W., Broach, E.R., Eds.; Cold Springs Harbor Laboratory Press: Cold Springs Harbor, NY, USA, 1981; pp. 211–287.
- Liti, G. The fascinating and secret wild life of the budding yeast S. cerevisiae. Elife 2015, 4, e05835.
- Krogerus, K.; Magalhães, F.; Vidgren, V.; Gibson, B. Novel brewing yeast hybrids: Creation and application. Appl. Microbiol. Biotechnol. 2017, 101, 65–78.
- Gunge, N.; Nakatomi, Y. Genetic mechanisms of rare matings of the yeast saccharomyces cerevisiae heterozygous for mating type. Genetics 1972, 70, 41–58.
- Ferenczy, L.; Maráz, A. Transfer of mitochondria by protoplast fusion in Saccharomyces cerevisiae. Nature 1977, 268, 524–525.
- Ogata, T.; Okumura, Y.; Tadenuma, M.; Tamura, G. Improving Transformation Method for Industrial Yeasts: Construction of ADH1-APT2 Gene and Using Electroporation. J. Gen. Appl. Microbiol. 1993, 294, 285–294.
- Alexander, W.G.; Peris, D.; Pfannenstiel, B.T.; Opulente, D.A.; Kuang, M.; Hittinger, C.T. Efficient engineering of marker-free synthetic allotetraploids of Saccharomyces. Fungal Genet. Biol. 2016, 89, 10–17.
- Strong, G.; England, K. Beer Style Guidelines—BJCP; Brewers Association: Boulder, CO, USA, 2021.
- Verstrepen, K.J.; Derdelinckx, G.; Dufour, J.P.; Winderickx, J.; Thevelein, J.M.; Pretorius, I.S.; Delvaux, F.R. Flavor-active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 2003, 96, 110–118.
- Mcmurrough, I.; Madigan, D.; Donnelly, D.; Hurley, J.; Doyle, A.; Hennigan, G.; McNulty, N. Control of Ferulic Acid and 4-Vinyl Guaiacol in Brewing. J. Inst. Brew. 1996, 102, 327–332.
- Krogerus, K.; Fletcher, E.; Rettberg, N.; Gibson, B.; Preiss, R. Efficient breeding of industrial brewing yeast strains using CRISPR/Cas9-aided mating-type switching. Appl. Microbiol. Biotechnol. 2021, 105, 8359–8376.
- Gibson, B.; Geertman, J.M.A.; Hittinger, C.T.; Krogerus, K.; Libkind, D.; Louis, E.J.; Magalhães, F.; Sampaio, J.P. New yeasts-new brews: Modern approaches to brewing yeast design and development. FEMS Yeast Res. 2017, 17, 1–13.
- Krogerus, K.; Preiss, R.; Gibson, B. A unique saccharomyces cerevisiae saccharomyces uvarum hybrid isolated from norwegian farmhouse beer: Characterization and reconstruction. Front. Microbiol. 2018, 9, 2253.
- Magalhães, F.; Krogerus, K.; Castillo, S.; Ortiz-Julien, A.; Dequin, S.; Gibson, B. Exploring the potential of Saccharomyces eubayanus as a parent for new interspecies hybrid strains in winemaking. FEMS Yeast Res. 2017, 17, 1–10.
- Giannakou, K.; Visinoni, F.; Zhang, P.; Nathoo, N.; Jones, P.; Cotterrell, M.; Vrhovsek, U.; Delneri, D. Biotechnological exploitation of Saccharomyces jurei and its hybrids in craft beer fermentation uncovers new aroma combinations. Food Microbiol. 2021, 100, 103838.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.2K
Revisions:
2 times
(View History)
Update Date:
22 Feb 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




