Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Shuiqiao Yuan | + 1824 word(s) | 1824 | 2022-02-16 07:11:08 | | | |
| 2 | Rita Xu | Meta information modification | 1824 | 2022-02-17 03:32:38 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Yuan, S. Spermatogenesis and Male Fertility. Encyclopedia. Available online: https://encyclopedia.pub/entry/19558 (accessed on 08 February 2026).
Yuan S. Spermatogenesis and Male Fertility. Encyclopedia. Available at: https://encyclopedia.pub/entry/19558. Accessed February 08, 2026.
Yuan, Shuiqiao. "Spermatogenesis and Male Fertility" Encyclopedia, https://encyclopedia.pub/entry/19558 (accessed February 08, 2026).
Yuan, S. (2022, February 17). Spermatogenesis and Male Fertility. In Encyclopedia. https://encyclopedia.pub/entry/19558
Yuan, Shuiqiao. "Spermatogenesis and Male Fertility." Encyclopedia. Web. 17 February, 2022.
Copy Citation
Spermatogenesis is an extremely complex developmental process and involves the orderly differentiation of multiple types of spermatogenic cells, including mitotically proliferating spermatogonial cells, meiotically dividing spermatocytes, and spermatids that eventually mature into spermatozoa.
mettl21a
methyltransferases
spermatogenesis
1. Introduction
In recent years, an increasing number of studies focused on the critical roles of epigenetic regulation in spermatogenesis [1]. Protein methylation, a post-translational modification, has been revealed to regulate a wide array of cellular functions ranging from RNA metabolism to chromatin structure remodeling, DNA repair, gene transcription, protein synthesis, and signal transduction [2].
With advances in mass spectrometry and characterization of the methylproteome, numerous protein methyltransferases (PMTs) were identified, which catalyze the methylation of histones and non-histone proteins [3]. Interestingly, many histone methyltransferases have been demonstrated their ability to regulate the development of germ cells at different stages of spermatogenesis. For instance, histone methyltransferase ESET can regulate the apoptosis of spermatogonial stem cells (SSC) by suppressing Cox4i2 expression by histone H3 lysine 9 tri-methylation (H3K9me3) [4]; combined depletion of EZH1 and EZH2, which are two protein methyltransferase that cause a depletion of global H3K27me3 marks and meiotic arrest in spermatocytes [5]; and conditional ablation of SETD2, a key methyltransferase catalyzing the trimethylation of histone H3 lysine 36 (H3K36me3), results in aberrant spermiogenesis with acrosomal malformation, leading to complete infertility [6].
Meanwhile, many methylated non-histone proteins and the enzymes that introduce them govern various biological processes [3], but their roles in spermatogenesis remain unclear. In humans, a group of methyltransferase-like 21 (METTL21) proteins (A–E) has recently been identified to mediate the lysine methylation of molecular chaperones and eukaryotic translation elongation factor 1A and closely relate with human health and disease [2][7][8][9][10][11]. Specifically, METTL21A catalyzes the methylation of HSP70 at Lys-561, which has been strongly linked to cancer and is often upregulated in tumors [12]; METTL21B could target Lys-165 in eEF1A in a GTP-dependent manner and has been described as one of the epigenetic factors for multiple sclerosis [8]; METTL21C, a protein-lysine N-methyltransferase catalyzing the Lys315 trimethylation in valosin-containing protein (VCP), can regulate calcium homeostasis in muscles and promote the differentiation of myoblasts to myotubes [9][10]. METTL21D, another VCP-methyltransferase, can promote tumor metastasis by affecting cell growth, migration, and infection [10][13].
2. Mettl21a Is Preferentially Expressed in Mouse Testes
Phylogenetic analyses and multi-alignment of METTL21A orthologs in 10 vertebrate species revealed that METTL21A is highly conserved during evolution (Figure 1a). To explore the expression profile of Mettl21a, researchers examined its mRNA and protein levels in multiple tissues from adult mice by RT-qPCR and Western blot analyses, respectively. The results showed that both mRNA and protein levels of Mettl21a are predominantly expressed in testis, followed by ovary, brain, and stomach (Figure 1b–d). They further investigated the spatial expression of Mettl21a during spermatogenesis and found that Mettl21a is continuously expressed from postnatal day 0 (P0) until P56 and displays the highest level at P56 (Figure 1e,f). These findings suggest a potential role of METTL21A in the regulation of spermatogenesis.
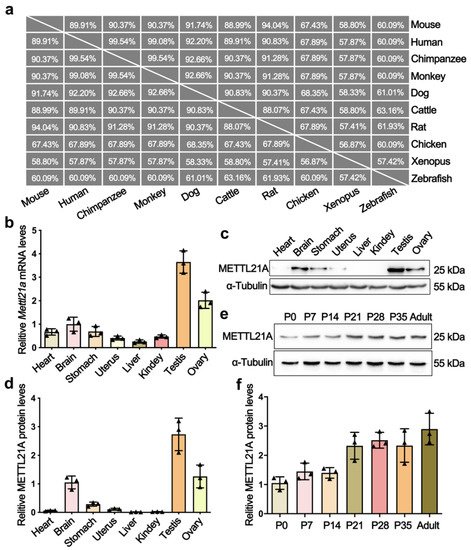
Figure 1. The expression profiles of METTL21A in multiple tissues and developing testes from mice. (a) A high degree of conservation of METTL21A in amino acid sequences was acquired from the NCBI database among 10 species. (b) RT-qPCR showing Mettl21a mRNA levels in multiple organs using Arbp as internal control. Data are presented as mean ± SEM, n = 3. The three values are indicated as three triangles on each bar. The following bar charts are the same unless otherwise stated. (c) Western blot is showing the protein levels of Mettl21a in various organs. (d) Quantification of relative Mettl21a protein levels in (c). (e) Western blot showing the protein levels of Mettl21a in developing testes. (f) Quantification of relative Mettl21a protein levels in (d). Data are presented as mean ± SEM, n = 3.
3. Generation of Mettl21a Knockout Mice
In order to elucidate the physiological role of METTL21A in spermatogenesis, researchers created Mettl21a knockout mouse model using CRISPR/cas9 technology. Two sgRNAs were, respectively, designed to target introns flanking the exon 2 (Figure 2a), thus generating a 1722-bp deletion verified by Sanger sequencing analysis, which resulted in frame-shift mutations of the Mettl21a gene. Specific primers for wild-type (WT) or homozygous mutant (KO) alleles were designed for genotyping (Figure 2b). RT-qPCR and Western blot confirmed the absence of both mRNA and protein of Mettl21a in KO testes (Figure 2c,d). Furthermore, by co-staining METTL21A and TRA98 (a nuclear marker of pan-germ cells), they found that METTL21A is mainly localized in the cytoplasm of germ cells, but it is absent in the KO testes (Figure 2e–f’’). Taken together, their data demonstrate that the Mettl21a gene was successfully knocked out in mice.
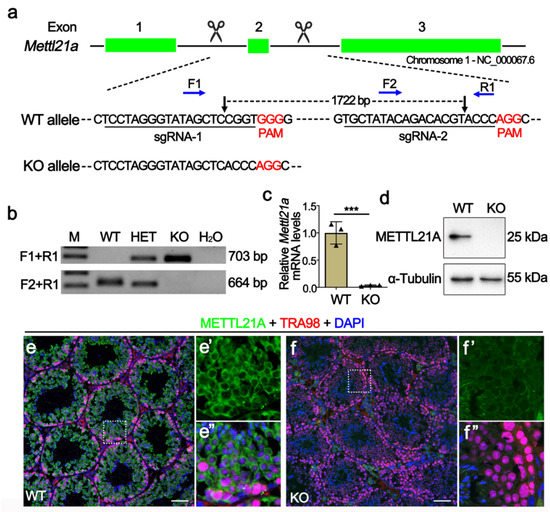
Figure 2. Generation of Mettl21a knockout mice using CRISPR/Cas9 technology. (a) Schematic representation of the targeting strategy for generating Mettl21a knockout (KO) mice using the CRISPR/Cas9 system. Green boxes represent exons of the Mettl21a gene on mouse chromosome 1; black underlines indicate the target regions of sgRNAs and black arrows show the cut sites, blue arrows indicate the sites of forward (F) and reverse (R) primers for genotyping. (b) Representative PCR genotyping results of Mettl21a alleles show that the WT allele is a longer band (703bp) using F1 and R1 primers, and the mutant allele is a shorter band (664bp) using F1 and R2 primers. M, marker; Het, heterozygous; H2O, negative control. (c) RT-qPCR analyses of Mettl21a mRNA levels in P56 WT and KO testes. Data are presented as mean ± SEM, n = 3. *** p < 0.001 by student’s t-test. (d) Western blot showing the METTL21A protein levels in WT and KO testes at P56. α-Tubulin served as a loading control. (e–f’’) Co-immunofluorescence staining of METTL21A and TRA98 (a nuclear marker of pan-germ cells) in WT and KO mouse testicular sections. Nuclei were stained with DAPI. Scale bars = 50 µm. Magnified images (e’,e’’,f’,f’’) of white boxes in (e) and (f), further highlight the changes in METTL21A localization and expression between WT and KO testes.
4. Mettl21a Is Dispensable for Spermatogenesis
Mettl21a KO mice were viable and developed normally, and fertility test by interbreeding adult WT or KO males with WT females for six months yielded no significant change in the number of litters and the average number of offspring per litter between WT and KO males (Figure 3a), indicating that Mettl21a KO males are completely fertile. Furthermore, adult Mettl21a KO males were indistinguishable from their WT littermates for testis size, testis/body weight ratio, the epididymal sperm count, and histological appearance of seminiferous tubule sections (Figure 3b–e). Seminiferous tubules from Mettl21a KO males harbor a full array of spermatogenic cells, including spermatogonia, spermatocytes, round spermatids, and elongated spermatids (Figure 3e). Consistent with these findings, a comparable number of mature spermatozoa was observed in the lumen of epididymis from WT and Mettl21a KO males (Figure 3f). These data suggest that spermatogenesis was not grossly impaired in the absence of Mettl21a in mice.
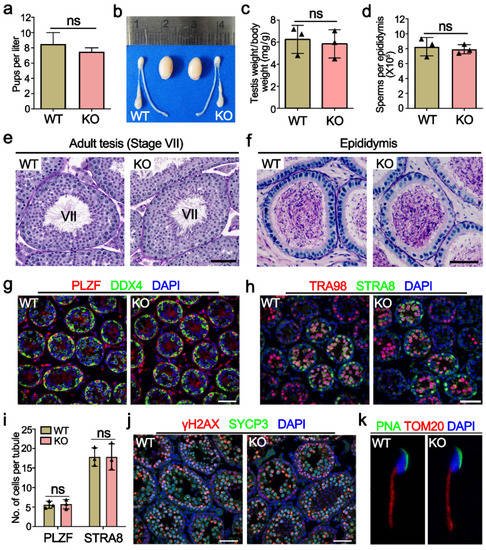
Figure 3. METTL21A is dispensable for spermatogenesis in mice. (a) The number of pups per litter derived from WT and KO males at the ages of 6–12 weeks. Data are presented as mean ± SEM, n = 8. ns, not significant. (b) The gross morphology of the testes from P56 WT and KO mice. (c) The ratio of testis weight to the body weight of 8-week-old WT and KO mice. Data are presented as mean ± SEM, n = 3. ns, not significant. (d) The sperm concentration of 8-week-old WT and KO mice. Data are presented as mean ± SEM, n = 3. ns, not significant. (e,f) Periodic acid-Schiff (PAS) staining of testes (e) and cauda epididymides (f) paraffin sections from 8-week-old WT and KO mice. Scale bar = 50 μm. (g) Co-immunofluorescence staining of PLZF and DDX4 in P10 WT and KO mouse testes. Scale bar = 50 µm. (h) Co-immunofluorescence staining of TRA98 and STRA8 in P10 WT and KO mouse testes. Scale bar = 50 µm. (i) The quantification of PLZF+ and STRA8+ cells per tubule at P10 in WT and KO testes, respectively. Data are presented as mean ± SEM, n = 3. ns, not significant. (j) Co-immunofluorescence staining of γH2AX and SYCP3 in P21 WT and KO testes. Scale bars = 50 µm. (k) Co-immunofluorescence staining of PNA and TOMM20 in spermatozoon from P56 WT and KO cauda epididymis.
In order to address more subtle defects, researchers immunostained several markers of germ cells and found no significant difference between WT and Mettl21a KO testis, including spermatogonia makers PLZF (undifferentiated spermatogonia maker) and STRA8 (a marker of differentiating spermatogonia and pre-leptotene spermatocytes) (Figure 3g–i); spermatocyte marker SYCP3 (a component of the axial elements of the synaptonemal complex); and DSB marker γH2AX (a phosphorylated form of the histone variant H2AX that accumulates upon formation of DSBs) (Figure 3j). Furthermore, Mettl21a KO spermatozoa display normal morphology labeled with acrosome marker PNA and mitochondrial marker TOM20 (labeling the midpiece of spermatozoa) (Figure 3k).
5. Deletion of Mettl21a Did Not Alter the Expression of Its Target Proteins and Other Mettl21 Members
Considering that Mettl21a can affect the expression and localization of its target proteins (HSP70, HSC70, GRP75, and GRP78) through methylation modification [14], researchers performed IF and Western blot assay to determine the localization and protein levels of its target proteins in Mettl21a KO testes. Similarly to the cellular distribution of METTL21A, these target proteins are also mainly located in the cytoplasm of germ cells and exhibit no apparent changes in Mettl21a KO testes compared with WT testes (Figure 4a–j). In addition, their protein levels are also indistinguishable between WT and Mettl21a KO testes (Figure 4k–l).
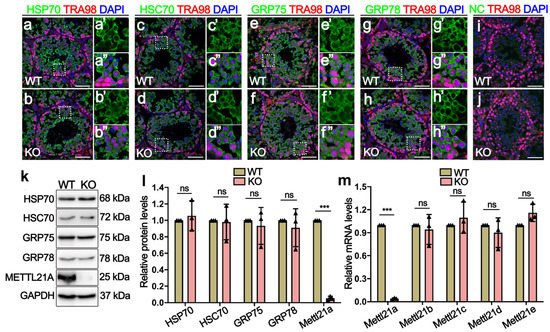
Figure 4. Mettl21a deletion does not affect the temporal and spatial expression of its target proteins. (a–h’’) Co-immunofluorescence staining of TRA98 and HSP70 (a,b), HSC70 (c,d), GRP75 (e,f), and GRP78 (g,h), respectively, in P56 WT and KO testes. Scale bar = 50 µm. Magnified images (a’,a’’,b’,b’’,c’,c’’,d’,d’’,e’,e’’,f’,f’’,g’,g’’,h’,h’’) of white boxes in (a–h), further highlight the expression and localization of HSP70, HSC70, GRP75, and GRP78 in WT and KO testes, respectively. (i,j) Negative control (NC) omitting the primary antibody to detect the specificity of the immunostaining with Alexa 488. Germ cells show unspecific staining. Germ cell were stained with anti-TRA98 antibody. Nuclei were stained with DAPI. Scale bars = 50 µm. (k) Western blot showing the expression of HSP70, HSC70, GRP75, GRP78, and METTL21A in P56 WT and KO testes. GAPDH was used as a loading control. (l) Quantification of relative indicated protein levels of B, respectively. Data are presented as mean ± SEM, n = 3. ns, not significant. *** p < 0.001 by student’s t-test. (m) RT-qPCR shows the mRNA levels of five members of the Mettl21 family in adult WT and KO testes. Data are presented as mean ± SEM, n = 3. ns, not significant. *** p < 0.001 by student’s t-test.
References
- McSwiggin, H.; O’Doherty, A. Epigenetic reprogramming during spermatogenesis and male factor infertility. Reproduction 2018, 156, R9–R21.
- Biggar, K.K.; Li, S.S.-C. Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 2015, 16, 5–17.
- Hwang, J.W.; Cho, Y.; Bae, G.-U.; Kim, S.-N.; Kim, Y.K. Protein arginine methyltransferases: Promising targets for cancer therapy. Exp. Mol. Med. 2021, 53, 788–808.
- Mahgoub, M.; Paiano, J.; Bruno, M.; Wu, W.; Pathuri, S.; Zhang, X.; Ralls, S.; Cheng, X.; Nussenzweig, A.; Macfarlan, T. Dual histone methyl reader ZCWPW1 facilitates repair of meiotic double strand breaks in male mice. Elife 2020, 9, e53360.
- An, J.; Zhang, X.; Qin, J.; Wan, Y.; Hu, Y.; Liu, T.; Li, J.; Dong, W.; Du, E.; Pan, C.; et al. The histone methyltransferase ESET is required for the survival of spermatogonial stem/progenitor cells in mice. Cell Death Dis. 2014, 5, e1196.
- Mu, W.; Starmer, J.; Shibata, Y.; Yee, D.; Magnuson, T. EZH1 in germ cells safeguards the function of PRC2 during spermatogenesis. Dev. Biol. 2017, 424, 198–207.
- Zuo, X.; Rong, B.; Li, L.; Lv, R.; Lan, F.; Tong, M.-H. The histone methyltransferase SETD2 is required for expression of acrosin-binding protein 1 and protamines and essential for spermiogenesis in mice. J. Biol. Chem. 2018, 293, 9188–9197.
- Cloutier, P.; Lavallée-Adam, M.; Faubert, D.; Blanchette, M.; Coulombe, B. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 2013, 9, e1003210.
- Małecki, J.; Aileni, V.K.; Ho, A.Y.; Schwarz, J.; Moen, A.; Sørensen, V.; Nilges, B.S.; Jakobsson, M.E.; Leidel, S.; Falnes, P.Ø. The novel lysine specific methyltransferase METTL21B affects mRNA translation through inducible and dynamic methylation of Lys-165 in human eukaryotic elongation factor 1 alpha (eEF1A). Nucleic Acids Res. 2017, 45, 4370–4389.
- Huang, J.; Hsu, Y.; Mo, C.; Abreu, E.; Kiel, D.P.; Bonewald, L.F.; Brotto, M.; Karasik, D. METTL21C is a potential pleiotropic gene for osteoporosis and sarcopenia acting through the modulation of the NF-κB signaling pathway. J. Bone Miner. Res. 2014, 29, 1531–1540.
- Kernstock, S.; Davydova, E.; Jakobsson, M.; Moen, A.; Pettersen, S.J.; Mælandsmo, G.M.; Egge-Jacobsen, W.; Falnes, P.Ø. Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat. Commun. 2012, 3, 1–11.
- Wang, C.; Zhang, B.; Ratliff, A.C.; Arlington, J.; Chen, J.; Xiong, Y.; Yue, F.; Nie, Y.; Hu, K.; Jin, W.; et al. Methyltransferase-like 21e inhibits 26S proteasome activity to facilitate hypertrophy of type IIb myofibers. FASEB J. 2019, 33, 9672–9684.
- Sieber, J.; Wieder, N.; Ostrosky-Frid, M.; Dvela-Levitt, M.; Aygün, O.; Udeshi, N.D.; Carr, S.A.; Greka, A. Lysine trimethylation regulates 78-kDa glucose-regulated protein proteostasis during endoplasmic reticulum stress. J. Biol. Chem. 2017, 292, 18878–18885.
- Moein-Vaziri, N.; Phillips, I.; Smith, S.; Almiňana, C.; Maside, C.; Gil, M.A.; Roca, J.; Martinez, E.A.; Holt, W.V.; Pockley, A.G.; et al. Heat-shock protein A8 restores sperm membrane integrity by increasing plasma membrane fluidity. Reproduction 2014, 147, 719–732.
More
Information
Subjects:
Reproductive Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Revisions:
2 times
(View History)
Update Date:
17 Feb 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




