Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Abu Hazafa | + 2344 word(s) | 2344 | 2022-02-11 03:11:45 | | | |
| 2 | Camila Xu | Meta information modification | 2344 | 2022-02-17 02:12:53 | | | | |
| 3 | Amina Yu | Meta information modification | 2344 | 2022-03-14 10:42:23 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Hazafa, A. Fungal Proteases. Encyclopedia. Available online: https://encyclopedia.pub/entry/19512 (accessed on 07 February 2026).
Hazafa A. Fungal Proteases. Encyclopedia. Available at: https://encyclopedia.pub/entry/19512. Accessed February 07, 2026.
Hazafa, Abu. "Fungal Proteases" Encyclopedia, https://encyclopedia.pub/entry/19512 (accessed February 07, 2026).
Hazafa, A. (2022, February 16). Fungal Proteases. In Encyclopedia. https://encyclopedia.pub/entry/19512
Hazafa, Abu. "Fungal Proteases." Encyclopedia. Web. 16 February, 2022.
Copy Citation
Proteases are obtained from plants, animals, and microorganisms. Plants and animals’ proteases are more complex as compared to microorganisms. Fungal proteases are used as detergents for the removal of stains by hydrolyzing the peptide bond among the protein molecules.
fungal enzymes
proteases
catalytical properties
1. Introduction
Excessive use of chemicals in different industries has increased tremendously in the past few years, including phenols, pesticides, polyaromatic hydrocarbons (PAHs), polychlorinated compounds, polychlorinated biphenyls, and arsenic. Exposure to these toxic chemicals has become a threat to the environment and public health concerns [1]. There is a need to replace these chemical compounds with fungal enzymes that are considered alternatives to the toxic chemicals and act as ecofriendly indicators to meet industrial demands [1]. The global market of fungal enzymes is drastically increasing in different sectors due to the high production rate, smooth downstream processing, and low costs [2][3].
Fungi are considered GRAS (Generally Regarded as Safe) organisms as compared to other microorganisms because they fulfill the criteria of industrial demands such as efficient growth on culture media in short duration and continuous supply of desired products [4][5]. Fungi also secreted a large variety of proteases, lipases, amylases, and amylases that play an important role in physiological processes such as germination, as defensins against other pathogens or for nutritional requirements for development [6][7]. Secretions of fungal enzymes occur from the cells present at the top of hyphae. These secreted enzymes can be used for industrial preparations of valuable products [8].
Fungal proteases have been widely studied due to their wide diversity [9]. Proteases have been isolated from different fungi such as Schizophyllum commune, Pleurotus ostreatus, Phanerochaete chrysosporium, Thermomyces lanuginosus, Sporotrichum thermophiles, Myceliophthora thermophile, Thermomyces ibadanensis, Candida mogii, Saccharomyces pombe, Aspergillus flavus, and Neurospora crassa [10][11][12]. Fungal proteases can be isolated through the fermentation process exhibiting high catalytic and specificity for the substrate [1][13].
Fungal proteases have diverse importance in corporate sectors such as the pharmaceutical, detergent, leather, waste, and food industries [14]. In the food industry, they are used to make beer, wine, and vinegar. Acidic proteases are used to improve wheat gluten’s structural and functional properties [15]. In medical sectors, fungal proteases are used as therapeutic agents for the treatment of a variety of diseases such as cancer, HIV, inflammatory diseases, diabetes, and hepatic cancer [16][17]. In the textile and laundry industries, they are used to prepare enzyme-based detergents to remove the tough stains from clothes due to developing excellent washing performance compared to other microbial enzymes [13][18]. They are also involved in degrading lignocellulosic biomass, and products can be utilized as biofuels for the production of energy at the commercial level [8][19].
2. Fungal Proteases
Proteases are obtained from plants, animals, and microorganisms. Plants and animals’ proteases are more complex as compared to microorganisms. Fungal proteases are used as detergents for the removal of stains by hydrolyzing the peptide bond among the protein molecules. Different strains of fungus produce the proteases, including Aspergillus niger [20]. Proteases have advantages over other fungus enzymes as detergents. Their demand increased due to low compatible detergent due to washing performance as compared to other enzymes. They catalyzed the removal of protein strains by breaking the peptide bonds in all pH ranges, such as acidic, neutral, and alkaline [21]. Their use in industrial and households increased due to the high production rate, enzyme recovery from the respective media, environmental safety, the whiteness of clothes, and maintenance of fibers.
Among these enzymes, protease is mainly used in detergent formulations due to its immense use in different industries. Proteases are referred to as proteolytic enzymes, which are unique in nature due to their presence in all living organisms and their significant applications in cell growth and differentiation. The proteases may be divided into three main groups based on the pH range alkaline protease, acidic protease, and neutral protease. Acidic protease has an optimum pH between 2.0 to 5.0. The ideal pH condition of neutral protease is 7.0, and alkaline protease has an optimum pH of more than 7.0. The acidic protease mainly has a fungus origin. Alkaline proteases are most important due to their immense applications in the food and detergent industries [22].
Protease enzymes are classified into two types based on their mechanism of action: extracellular protease and intracellular protease. The extracellular enzyme catalyses the hydrolysis of large protein molecules into smaller molecules that are absorbed into the cells. The intracellular protease plays a significant role in the regulation of metabolism. The extracellular protease enzyme is mostly used at a commercial scale to degrade protein molecules in many industrial processes [23]. The structures of different proteases are presented in Figure 1.
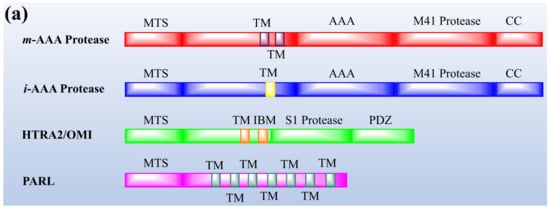
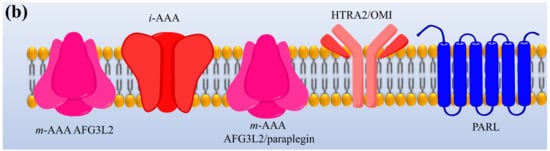
Figure 1. The representation of (a) structure and (b) topology of proteases. TM: Transmembrane domain, IBM: Inhibitor of apoptosis (IAP)-binding motif, MTS: Mitochondrial targeting sequencing, IMS: Intermembrane space, CC: Coiled-coil, AAA: Triple-A domain, M41: Protease metal-binding proteolytic domain, S1 protease: Trypsin-like protease domain. This figure is reproduced from Martinelli and Rugarli [24] after permission from Elsevier (License No. 5197711293142).
Alkaline proteases are used as detergents in the laundry industry to remove the dirty stains that are portentous in nature, in many food industries such as cheese making, baking meat industries, soaking processes, and many others in waste management from many food industries and also have some application in household activities [25]. Alkaline proteases act as an active ingredient in laundry detergent; they are considered a significant application of this enzyme. In the past, when detergent protease was produced at an industrial scale, it caused some allergic problems in workers from the dust of enzymes [26].
Among all groups or classes of proteases, the serine proteases are most effective in detergent action due to their stability and comparability in the existence of ingredients and other bleaching agents. It is expected that in 2020, the global market of industrial reach will climb to 7.5 billion and the growth rate will be around 8.2%. It is estimated that the growth rate will be maximum in detergents’ enzymes. One type of such enzyme, protease, remained a well-known enzyme of 2015; its growth rate was 27.5%. It is expected to enhance its growth rate because of its flexibility in different sectors of pharmaceutical foods and detergents [27]. The information about novel protease enzymes isolated from different sources is presented in Table 1.
Table 1. The representation of novel fungal protease enzymes isolated from different sources.
| Enzyme Isolated | Enzyme Class | Active Site Residue (s) | Isolated Source | Reference |
|---|---|---|---|---|
| Clostripain, Streptopain | Cysteine proteases | Cysteine and histidine residues | C. histolyticum, S. griseus | [28][29] |
| Pepsins, proteases, rennet like proteases | Aspartic endoproteases | Two aspartate residues | A. niger, M. miehei | [30][31] |
| Chymotrypsins, subtilisins | Serine proteases | Serine residues | B. sphaericus | [32][33] |
| Collagenases, elastase | Metalloendoproteases | Metal ions | C. histolyticum, P. aeruginosa |
[34][35] |
| Eqolisin protease | Glutamic proteases | Glutamate residues | S. lignicola, A. niger | [36][37] |
| Pepsins (A1), retropepsin (A2) | Acidic proteases | - | A. niger, A. saitoi. | [38][39] |
| Subtilisin, carlsberg | Alkaline proteases | - | A. salinivibrio, C. aureus | [39] |
| Neutrase, thermolysin | Neutral proteases | - | Bacillus sp. | [39] |
2.1. Origin of Fungal Proteases
The demand for fungal proteases has increased in recent decades. Different species of fungi secrete proteases, but the origin of proteases from basidiomycetes remains unclear [40]. Aspergillus species are considered an excellent source of proteases. Some other fungal species such as Penicillium and Rhizopus also produce proteases [41][42]. Proteases are also produced from the basidiomycetes such as Schizophyllum commune, Armillariella mellea, Pleurotus ostreatus, and Phanerochaete chrysosporium [43][44][45].
Mycelial secretion in saprophytic basidiomycetes led to discovery of proteases such as subtilases from Serpula lacrymans, Pleurotus ostreatus, and Irpex lacteus [46][47]. Pleurotus sp., such as P. ostreatus and P. chrysosporium, also produced the proteases involved in ligninolytic mechanism through fragmented degradation of laccase enzyme during fugal growth [48]. Hemolytic proteases secreted by Pleurotus sp. possess different activities in cellular processes. Secretion of hemolysin by P. nebrodensis shows apoptosis and antiproliferative activities and is involved in targeting cancerous cells [49]. These proteases tightly bind to the receptor proteins of HIV and inhibit them [50]. Some proteases isolated from P. ostreatus possess activities against different carcinomas [51].
2.2. Classification of Fungal Proteases
Fungal proteases are classified into different categories based on amino acids. A few are presented in the following sections.
2.2.1. Protease Classes Based on Amino Acids
Proteases are categorized into the following classes based on amino acids’ residue in their active site.
Serine Protease
Serine proteases are the most important class of proteases that contain the amino acid serine in their active site (see Figure 2). Serine residues in the active site make a catalytic triad with aspartate and histidine. This catalytic triad is conserved among all serine proteases. These residues are essential for their catalytic activity to cleave the substrate protein [52]. Serine proteases can be further divided based on substrate specificities such as elastase-like proteases, which possess the smaller S1 cleft than the chymotrypsin-like protease. They are hydrophobic in nature and possess the specificity for valine and glycine. These elastase-like proteases act on elastin by breaking them into smaller fragments, thus playing an essential role in forming connective tissues [3]. Fungal species such as Aspergillus sp., Aspergillus oryzae, Aspergillus fischeri, Penicillium citrinum, Penicillium corylophilum. Penicillium waksmanii, and Neurospora Conidiobol produce serine proteases [53][54].
Threonine Protease
Threonine proteases hold the threonine residues in their active site for their catalytic activity to cleave the substrate protein. These proteases show substrate specificity for bulky amino acids [39][55]. Saccharomyces cerevisiae are production sources of threonine peptidase with appreciable capacity for the production of threonine and potential for industrial application [53][56].
Cysteine Protease
Cysteine proteases hold the two amino residues in their active sites, such as cysteine and histidine (see Figure 2). Their catalytic activity can be maintained in the presence of reducing agents [39]. The substrate specificity of the cysteine proteases can be determined through the interaction between the side-chain amino acids of the particular substrate that can be accommodated into the S2 cleft, which is hydrophobic in nature and shows the specificity for the leucine and tyrosine. One of the best examples is the Cathepsin K that shows the specificity for kinins by cleaving the peptide bonds in the collagen tissue [28][57]. Fungi species such as Aspergillus oryzae produce cysteine proteases. Only a little information about the secretion of fungal cysteine proteases is known [58].
Aspartate Protease
Aspartate proteases hold the two aspartate residues in their active site. These aspartate residues are essential for their catalytic activity to cleave the substrate protein (see Figure 2). Aspartate proteases show specificity for aromatics such as phenylalanine, tyrosine, and tryptophan on both sides of the peptide bond [31]. Rennin-like aspartate proteases cause the cleavage of casein into the smaller peptides. Pepsins are isolated from Aspergillus, and rennet-like enzymes are isolated from M. miehei [30]. Fungi species such as Aspergillus, Penicillium, Rhizopus, and Neurospora produce aspartic proteases [30][59]. E. parasitica and R. miehei of fungi are used as production sources of aspartic peptidase with appreciable capacity for the production of acidic peptidase and potential for industrial application [59][60].
Glutamic Acid Protease
Glutamic acid protease holds the glutamate residues in their active site. These proteases also show substrate specificity for bulky amino acids. For instance, eqolisin can work at pH 2.0 when the casein is used as a substrate. Glutamic proteases were identified in the fungi Scytalidium lignicola and Aspergillus niger [36][37]. Scytalidium lignicolum, Aspergillus niger, Cryphonectria parasitica, Talaromyces emersonii and Sclerotina sclerotiorum secreted glutamic proteases [61][62][63].
Metalloprotease
Metalloproteases are the diverse classes of proteases containing metal ions in their active sites (see Figure 2). These metalloproteases are highly specific in their action [34]. Neutral proteases show the specificity for hydrophobic amino acids. The well-known metalloproteases such as collagenases and elastase are isolated from C. histolyticum and P. aeruginosa [35]. Fungi species such as Aspergillus, Penicillium, Fusarium oxysporum, and A. fumigatus produce metalloproteases [30][56][64].
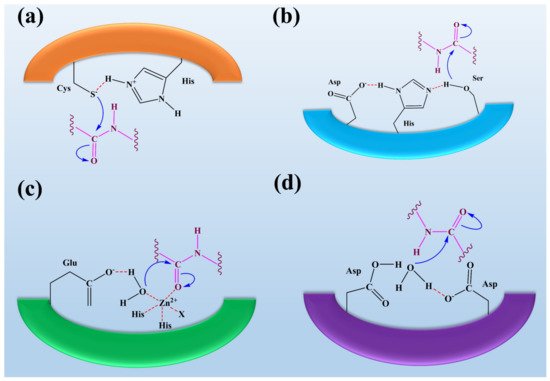
Figure 2. The representation of fungal protease mechanisms of (a) serine proteases (b) aspartyl proteases, (c) metalloproteases, and (d) cysteine proteases. It was reported that the eponymous residue is commonly formed as a pair with a proton withdrawing group in the active sites of cysteine and serine proteases to promote a nucleophilic attack on the peptide bond. In contrast, metalloproteases and aspartyl proteases activate water molecules as nucleophiles. Overall, it was observed that the process of peptide bond scission is the same for all classes of proteases. This figure is reproduced from Erez et al. [65] after permission from Springer Nature (License No. 5223461087907).
The mechanism of action of fungal proteases involves the formation of an intermediate acyl-enzyme that covalently links the enzyme to the N-terminal of the substrate. In the second step, the water molecule completes the hydrolysis by attacking the intermediate. It results in the release of the second-half product with the regeneration of free enzyme (see Figure 3) [66].
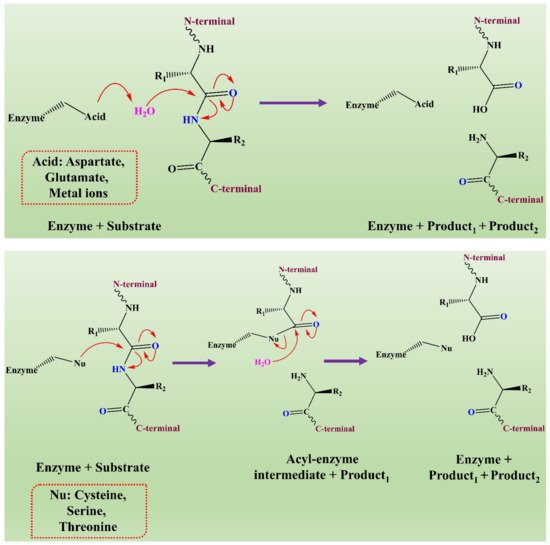
Figure 3. The representation of a comparison of the two hydrolytic mechanisms used for proteolysis. This figure is reproduced from Shafee [66] (Attribution NonCommercial 2.0 UK: England & Wales, CC BY-NY 2.0 UK).
3. Market Value of Fungal Proteases
Fungal proteases are one of the largest groups of industrial enzymes, and their global market is drastically increasing annually. Of the 60% of enzymes marketed worldwide, fugal proteases account for 20% [67][68]. The global market of fungal proteases is gaining more attention compared to other enzymes due to their high demand, catalytic properties, and low cost [69][70]. The global fungal enzymes market is highly consolidated owing to the presence of several key players operating in the global fungal enzymes market, namely Novozymes A/S, DSM, Chr. Hansen, DuPont, and BASF. The share market is divided into pharmaceuticals, bakery, beverages, sweet, and animal feed based on application [71]. Companies worldwide adopt evolutionary acquisition strategies by expanding the business network of proteases by considering the geographical demands, source type (especially microbial), and role in different industries [70].
Global Proteases Market Segmentation
The global protease market is segmented based on applications, forms, and types. The market is divided into renin, trypsin, pepsin, and others based on type. The renin segment leads the share market, and alkaline protease is the second segment that accounts for the largest market share due to usage in the food and dairy industry [72]. Based on form, the protease market has been divided into liquid and powder. The powder segments contribute to the largest market share due to the usage of many products that improve the half-life of food products [20]. The share market is divided into bakery and beverages, sweet, animal feed, and others based on application. The bakery segment contributes to the largest share of the market. It reflects the use of proteases in different industries for syntheses of food-based products [73].
References
- Monod, M.; Capoccia, S.; Léchenne, B.; Zaugg, C.; Holdom, M.; Jousson, O. Secreted proteases from pathogenic fungi. Int. J. Med. Microbiol. 2002, 292, 405–419.
- Singh, R.; Mittal, A.; Kumar, M.; Mehta, P.K. Microbial proteases in commercial applications. J. Pharm. Chem. Biol. Sci. 2016, 4, 365–374.
- Rao, M.B.; Tanksale, A.M.; Ghatge, M.S.; Deshpande, V.V. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 1998, 62, 597–635.
- Singh, N.; Gaur, S. GRAS Fungi: A New Horizon in Safer Food Product. In Fungi in Sustainable Food Production; Springer: Cham, Switzerland, 2021; pp. 27–37.
- Novelli, P.K.; Barros, M.M.; Fleuri, L.F. Novel inexpensive fungi proteases: Production by solid state fermentation and characterization. Food Chem. 2016, 198, 119–124.
- Souza, P.M.d.; Bittencourt, M.L.d.A.; Caprara, C.C.; Freitas, M.d.; Almeida, R.P.C.d.; Silveira, D.; Fonseca, Y.M.; Ferreira, E.X.; Pessoa, A.; Magalhães, P.O. A biotechnology perspective of fungal proteases. Braz. J. Microbiol. 2015, 46, 337–346.
- Kumar, A.; Gautam, A.; Dutt, D. Screening of fungal resources for the production of cellulases and xylanases. Biotechnol. J. Int. 2015, 9, 1–13.
- Lange, L. Fungal enzymes and yeasts for conversion of plant biomass to bioenergy and high-value products. Microbiol. Spectr. 2017, 5.
- Damare, S.; Raghukumar, C.; Muraleedharan, U.D.; Raghukumar, S. Deep-sea fungi as a source of alkaline and cold-tolerant proteases. Enzym. Microb. Technol. 2006, 39, 172–181.
- Kumar, A. Aspergillus nidulans: A Potential Resource of the Production of the Native and Heterologous Enzymes for Industrial Applications. Int. J. Microbiol. 2020, 2020, 8894215.
- Gnanadoss, J.J.; Devi, S.K. Optimization of nutritional and culture conditions for improved protease production by Aspergillus nidulans and Aspergillus flavus. J. Microbiol. Biotechnol. Food Sci. 2021, 2021, 518–523.
- Hoffmeister, D.; Keller, N.P. Natural products of filamentous fungi: Enzymes, genes, and their regulation. Nat. Prod. Rep. 2007, 24, 393–416.
- Savitha, S.; Sadhasivam, S.; Swaminathan, K.; Lin, F.H. Fungal protease: Production, purification and compatibility with laundry detergents and their wash performance. J. Taiwan Inst. Chem. Eng. 2011, 42, 298–304.
- Liu, E.; Li, M.; Abdella, A.; Wilkins, M.R. Development of a cost-effective medium for submerged production of fungal aryl alcohol oxidase using a genetically modified Aspergillus nidulans strain. Bioresour. Technol. 2020, 305, 123038.
- Sharma, M.; Gat, Y.; Arya, S.; Kumar, V.; Panghal, A.; Kumar, A. A review on microbial alkaline protease: An essential tool for various industrial approaches. Ind. Biotechnol. 2019, 15, 69–78.
- Sabotič, J.; Kos, J. Microbial and fungal protease inhibitors—current and potential applications. Appl. Microbiol. Biotechnol. 2012, 93, 1351–1375.
- Agbowuro, A.A.; Huston, W.M.; Gamble, A.B.; Tyndall, J.D. Proteases and protease inhibitors in infectious diseases. Med. Res. Rev. 2018, 38, 1295–1331.
- Bezerra, V.H.S.; Cardoso, S.L.; Fonseca-Bazzo, Y.; Silveira, D.; Magalhães, P.O.; Souza, P.M. Protease Produced by Endophytic Fungi: A Systematic Review. Molecules 2021, 26, 7062.
- Gupta, V.K.; Kubicek, C.P.; Berrin, J.-G.; Wilson, D.W.; Couturier, M.; Berlin, A.; Edivaldo Filho, X.; Ezeji, T. Fungal enzymes for bio-products from sustainable and waste biomass. Trends Biochem. Sci. 2016, 41, 633–645.
- Naveed, M.; Nadeem, F.; Mehmood, T.; Bilal, M.; Anwar, Z.; Amjad, F. Protease—a versatile and ecofriendly biocatalyst with multi-industrial applications: An updated review. Catal. Lett. 2021, 151, 307–323.
- El-Khonezy, M.I.; Elgammal, E.W.; Ahmed, E.F.; Abd-Elaziz, A.M. Detergent stable thiol-dependant alkaline protease produced from the endophytic fungus Aspergillus ochraceus BT21: Purification and kinetics. Biocatal. Agric. Biotechnol. 2021, 35, 102046.
- Niyonzima, F.N.; More, S.S. Screening and optimization of cultural parameters for an alkaline protease production by Aspergillus terreus gr. under submerged fermentation. Int. J. Pharm. Bio. Sci. 2013, 4, 1016–1028.
- Yadav, V.K.; Singh, V.; Mishra, V. Alkaline Protease: A Tool to Manage Solid Waste and Its Utility in Detergent Industry. In Microbial Genomics in Sustainable Agroecosystems; Springer: Berlin/Heidelberg, Germany, 2019; pp. 231–254.
- Martinelli, P.; Rugarli, E.I. Emerging roles of mitochondrial proteases in neurodegeneration. Biochim. Et Biophys. Acta (BBA)-Bioenerg. 2010, 1797, 1–10.
- Hariharan, A.; Rajadurai, U.M.; Palanivel, I. Isolation, Purification and Mass Production of Protease from Bacillus subtilis. 2019. Available online: https://ssrn.com/abstract=3370124 (accessed on 15 July 2021).
- Devi, M.K.; Banu, A.R.; Gnanaprabhal, G.; Pradeep, B.; Palaniswamy, M. Purification, characterization of alkaline protease enzyme from native isolate Aspergillus niger and its compatibility with commercial detergents. Indian J. Sci. Technol. 2008, 1, 1–6.
- Shahid, T.; Muhammad, S.; Ahmed, K. Enzyme Proteases Used in Laundry Detergents Engineering a Review. Sci. Int. 2016, 28, 2711–2717.
- Corvo, I.; Ferraro, F.; Merlino, A.; Zuberbühler, K.; O’Donoghue, A.J.; Pastro, L.; Pi-Denis, N.; Basika, T.; Roche, L.; McKerrow, J.H. Substrate specificity of cysteine proteases beyond the S2 Pocket: Mutagenesis and molecular dynamics investigation of Fasciola hepatica Cathepsins L. Front. Mol. Biosci. 2018, 5, 40.
- Ou, J.-F.; Zhu, M.-J. An overview of current and novel approaches for microbial neutral protease improvement. Int. J. Mod. Biol. Med 2012, 2, 1–31.
- Theron, L.W.; Divol, B. Microbial aspartic proteases: Current and potential applications in industry. Appl. Microbiol. Biotechnol. 2014, 98, 8853–8868.
- Dunn, B.M. Introduction to the aspartic proteinase family. Aspartic Acid Proteases Ther. Targets 2010, 45, 1–21.
- Jiang, L.; Zhang, X.; Zhou, Y.; Chen, Y.; Luo, Z.; Li, J.; Yuan, C.; Huang, M. Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases. RSC Adv. 2018, 8, 28189–28197.
- Jiang, L.; Yuan, C.; Huang, M. A general strategy to inhibit serine protease by targeting its autolysis loop. FASEB J. 2021, 35, e21259.
- Rawlings, N.D.; Barrett, A.J. Evolutionary families of metallopeptidases. Methods Enzymol. 1995, 248, 183–228.
- Cheng, M.; Takenaka, S.; Aoki, S.; Murakami, S.; Aoki, K. Purification and characterization of an eggshell membrane decomposing protease from Pseudomonas aeruginosa strain ME-4. J. Biosci. Bioeng. 2009, 107, 373–378.
- Sims, A.H.; Dunn-Coleman, N.S.; Robson, G.D.; Oliver, S.G. Glutamic protease distribution is limited to filamentous fungi. FEMS Microbiol. Lett. 2004, 239, 95–101.
- Kuan, C.S.; Ng, K.P.; Yew, S.M.; Meleh, H.U.; Seow, H.F.; How, K.N.; Yeo, S.K.; Jee, J.M.; Tan, Y.-C.; Yee, W.-Y. Comparative Genomic and Phenotypic Analyses of Pathogenic Fungi Neoscytalidium Dimidiatum and Bipolaris Papendorfii, Isolated From Human Skin Scraping. Res. Sq. 2020, 1–32.
- Mamo, J.; Assefa, F. The role of microbial aspartic protease enzyme in food and beverage industries. J. Food Qual. 2018, 2018, 7957269.
- Razzaq, A.; Shamsi, S.; Ali, A.; Ali, Q.; Sajjad, M.; Malik, A.; Ashraf, M. Microbial proteases applications. Front. Bioeng. Biotechnol. 2019, 7, 110.
- Erjavec, J.; Kos, J.; Ravnikar, M.; Dreo, T.; Sabotič, J. Proteins of higher fungi–from forest to application. Trends Biotechnol. 2012, 30, 259–273.
- Shaba, A.; Baba, J. Screening of Pleurotus ostreatus and Gleophylum sepiarium strains for extracellular protease enzyme production. Bayero J. Pure Appl. Sci. 2012, 5, 187–190.
- Papagianni, M. Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol. Adv. 2004, 22, 189–259.
- Wang, H.; Ng, T. Pleureryn, a novel protease from fresh fruiting bodies of the edible mushroom Pleurotus eryngii. Biochem. Biophys. Res. Commun. 2001, 289, 750–755.
- Musatti, A.; Ficara, E.; Mapelli, C.; Sambusiti, C.; Rollini, M. Use of solid digestate for lignocellulolytic enzymes production through submerged fungal fermentation. J. Environ. Manag. 2017, 199, 1–6.
- Burdsall, H.H., Jr.; Volk, T.J. Armillaria solidipes, an older name for the fungus called Armillaria ostoyae. North Am. Fungi 2008, 3, 261–267.
- Faraco, V.; Palmieri, G.; Festa, G.; Monti, M.; Sannia, G.; Giardina, P. A new subfamily of fungal subtilases: Structural and functional analysis of a Pleurotus ostreatus member. Microbiology 2005, 151, 457–466.
- Nurika, I.; Suhartini, S.; Barker, G.C. Biotransformation of tropical lignocellulosic feedstock using the brown rot fungus Serpula lacrymans. Waste Biomass Valorization 2020, 11, 2689–2700.
- Cha, W.-S.; Park, S.-S.; Kim, S.-J.; Choi, D. Biochemical and enzymatic properties of a fibrinolytic enzyme from Pleurotus eryngii cultivated under solid-state conditions using corn cob. Bioresour. Technol. 2010, 101, 6475–6481.
- Ng, T.B.; Wong, J.H.; Cheung, R.C.F.; Tse, T.F.; Tam, T.; Chan, H. Mushrooms: Proteins, polysaccharidepeptide complexes and polysaccharides with antiproliferative and anticancer activities. Int. J. Cancer Res. Prev. 2014, 7, 287.
- Lv, H.; Kong, Y.; Yao, Q.; Zhang, B.; Leng, F.-w.; Bian, H.-j.; Balzarini, J.; Van Damme, E.; Bao, J.-k. Nebrodeolysin, a novel hemolytic protein from mushroom Pleurotus nebrodensis with apoptosis-inducing and anti-HIV-1 effects. Phytomedicine 2009, 16, 198–205.
- Berne, S.; Križaj, I.; Pohleven, F.; Turk, T.; Maček, P.; Sepčić, K. Pleurotus and Agrocybe hemolysins, new proteins hypothetically involved in fungal fruiting. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2002, 1570, 153–159.
- Sumantha, A.; Larroche, C.; Pandey, A. Microbiology and industrial biotechnology of food-grade proteases: A perspective. Food Technol. Biotechnol. 2006, 44, 211.
- Sandhya, C.; Nampoothiri, K.M.; Pandey, A. Microbial Proteases. In Microbial Enzymes and Biotransformations; Springer: Berlin/Heidelberg, Germany, 2005; pp. 165–179.
- dos Santos Aguilar, J.G.; Sato, H.H. Microbial proteases: Production and application in obtaining protein hydrolysates. Food Res. Int. 2018, 103, 253–262.
- Baird, T.T., Jr.; Wright, W.D.; Craik, C.S. Conversion of trypsin to a functional threonine protease. Protein Sci. 2006, 15, 1229–1238.
- Jashni, M.K.; Dols, I.H.; Iida, Y.; Boeren, S.; Beenen, H.G.; Mehrabi, R.; Collemare, J.; de Wit, P.J. Synergistic action of a metalloprotease and a serine protease from Fusarium oxysporum f. sp. lycopersici cleaves chitin-binding tomato chitinases, reduces their antifungal activity, and enhances fungal virulence. Mol. Plant-Microbe Interact. 2015, 28, 996–1008.
- Barrett, A.J. Classification of peptidases. Methods Enzymol. 1994, 244, 1–15.
- Brocklehurst, K.; Philpott, M.P. Cysteine proteases: Mode of action and role in epidermal differentiation. Cell Tissue Res. 2013, 351, 237–244.
- Yegin, S.; Fernandez-Lahore, M.; Salgado, A.J.G.; Guvenc, U.; Goksungur, Y.; Tari, C. Aspartic proteinases from Mucor spp. in cheese manufacturing. Appl. Microbiol. Biotechnol. 2011, 89, 949–960.
- Da Silva, R.R.; Souto, T.B.; de Oliveira, T.B.; de Oliveira, L.C.G.; Karcher, D.; Juliano, M.A.; Juliano, L.; de Oliveira, A.H.; Rodrigues, A.; Rosa, J.C. Evaluation of the catalytic specificity, biochemical properties, and milk clotting abilities of an aspartic peptidase from Rhizomucor miehei. J. Ind. Microbiol. Biotechnol. 2016, 43, 1059–1069.
- Dietrich, F.S.; Voegeli, S.; Brachat, S.; Lerch, A.; Gates, K.; Steiner, S.; Mohr, C.; Pöhlmann, R.; Luedi, P.; Choi, S. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 2004, 304, 304–307.
- Feldmann, H. Génolevures—A Novel Approach to ‘Evolutionary Genomics’; Elsevier: Amsterdam, The Netherlands, 2000; Volume 487, pp. 1–2.
- Kellis, M.; Birren, B.W.; Lander, E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 2004, 428, 617–624.
- Neto, Y.A.A.H.; de Souza Motta, C.M.; Cabral, H. Optimization of metalloprotease production by Eupenicillium javanicum in both solid state and submerged bioprocesses. Afr. J. Biochem. Res. 2013, 7, 146–157.
- Erez, E.; Fass, D.; Bibi, E. How intramembrane proteases bury hydrolytic reactions in the membrane. Nature 2009, 459, 371–378.
- Shafee, T. Evolvability of a Viral Protease: Experimental Evolution of Catalysis, Robustness and Specificity; University of Cambridge: Cambridge, UK, 2014.
- Madhavan, A.; Arun, K.; Binod, P.; Sirohi, R.; Tarafdar, A.; Reshmy, R.; Awasthi, M.K.; Sindhu, R. Design of novel enzyme biocatalysts for industrial bioprocess: Harnessing the power of protein engineering, high throughput screening and synthetic biology. Bioresour. Technol. 2021, 325, 124617.
- Veloorvalappil, N.J.; Robinson, B.S.; Selvanesan, P.; Sasidharan, S.; Kizhakkepawothail, N.U.; Sreedharan, S.; Prakasan, P.; Moolakkariyil, S.J.; Sailas, B. Versatility of microbial proteases. Adv. Enzym. Res. 2013, 2013, 36957.
- Shankar, R.; Upadhyay, P.K.; Kumar, M. Protease Enzymes: Highlights on Potential of Proteases as Therapeutics Agents. Int. J. Pept. Res. Ther. 2021, 27, 1281–1296.
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial applications of enzymes: Recent advances, techniques, and outlooks. Catalysts 2018, 8, 238.
- Timson, D.J. Four challenges for better biocatalysts. Fermentation 2019, 5, 39.
- Banerjee, G.; Ray, A.K. Impact of microbial proteases on biotechnological industries. Biotechnol. Genet. Eng. Rev. 2017, 33, 119–143.
- Ademosun, M.T.; Omoba, O.S.; Olagunju, A.I. Antioxidant properties, glycemic indices, and carbohydrate hydrolyzing enzymes activities of formulated ginger-based fruit drinks. J. Food Biochem. 2021, 45, e13324.
More
Information
Subjects:
Microbiology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.8K
Revisions:
3 times
(View History)
Update Date:
14 Mar 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




