Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Wu, Y. T Cell Epitopes from the Proteome of HBV. Encyclopedia. Available online: https://encyclopedia.pub/entry/19440 (accessed on 11 March 2026).
Wu Y. T Cell Epitopes from the Proteome of HBV. Encyclopedia. Available at: https://encyclopedia.pub/entry/19440. Accessed March 11, 2026.
Wu, Yandan. "T Cell Epitopes from the Proteome of HBV" Encyclopedia, https://encyclopedia.pub/entry/19440 (accessed March 11, 2026).
Wu, Y. (2022, February 15). T Cell Epitopes from the Proteome of HBV. In Encyclopedia. https://encyclopedia.pub/entry/19440
Wu, Yandan. "T Cell Epitopes from the Proteome of HBV." Encyclopedia. Web. 15 February, 2022.
Copy Citation
T cell epitopes functionally validated from HBV antigens during the past 33 years; the human leukocyte antigen (HLA) supertypes to present these epitopes, and the methods to screen and identify T cell epitopes.
hepatitis B virus
T cell epitope
HLA restriction
1. Introduction
Hepatitis B virus (HBV) infection still disseminates across the world and causes the most common and fatal liver diseases including acute liver failure, chronic hepatitis, liver cirrhosis (LC), and hepatocellular carcinoma (HCC) [1][2]. Nucleoside analogs and/or interferon are widely utilized antiviral drugs, which can effectively suppress virus replication, decrease serum HBV DNA to undetectable levels, mitigate liver fibrosis, and reduce HCC risk [3][4][5], however cannot eliminate the virus in patients. Recurrence after therapy discontinuation is emerging to be a common etiology of morbidity and mortality in patients with chronic HBV infection[6] .
Numerous researches have demonstrated the important influence of HBV-specific T cell responses on virus clearance[7], disease progression[8][9][10], antiviral efficacy[11][12], and recurrence[13][14][15], particularly the CD8 T cells, which act as vital effector cells to kill virus-infected hepatocytes and secret cytokines. Patients with acute-resolving HBV infection show robust HBV-specific CD8 T cell responses, while the patients with chronic HBV infection present a phenomenon termed CD8 T cell functional exhaustion with multifactorial heterogeneity[9], and differs depending on the targeted antigen for HLA-A02 restricted epitopes located in the core antigen versus polymerase[16]. Furthermore, the heterogeneity of HBV-specific T cells also responds differently to therapeutic stimuli. Therefore, T cells specific for HBV not only are the potential markers for monitoring the effects of antiviral therapy and predicting the recurrence [17], but also are the promising modulators in specific immunotherapy. Identifying the T cell epitopes as many as possible from HBV antigens will greatly contribute to the design and development of epitope-based and T cell-based therapies and the detection of host HBV-specific T cell immunity. Although a systematic review of T cell epitopes in HBV antigens was reported in 2008[18], an updated map of the T cell epitopes is urgently needed.+++
2. HBV Proteome and the Approaches Identifying T Cell Epitopes
HBV is one of the smallest viruses with a genome length of 3.2 Kb [19]. Its genome contains four open reading frames (ORFs) coding four partially overlapping proteins as displayed in Figure 1: (1) preS/S ORF encodes large (L), middle (M), and small (S) surface antigens (HBsAg). HBsAg is being widely investigated in clinical fields and quantified as a diagnostic marker of HBV infection as it can reflect the level of covalently closed circular DNA (cccDNA) and intrahepatic HBV DNA in chronic infection[20][21]. (2) Pre-core/core ORF encodes hepatitis B e antigen (HBeAg), core antigen (HBcAg) or in combination core-related antigen (HBcrAg). HBeAg has long been advocated as a serum marker for guiding the clinical practice of chronic hepatitis B virus [22][23]. HBcrAg has been demonstrated more recently as a potential surrogate marker of cccDNA[24]. (3) X ORF encodes HBx antigen (HBxAg), which plays an important role in virus genome transcription and is correlated with liver cancer. The expression of HBxAg in HBV-associated HCC patients is significantly higher than other viral proteins [25]. (4) P ORF encodes the viral DNA polymerase (HBpol), which is responsible for the replication of the viral genome and is an effective target for the therapeutic intervention of chronic HBV infection [26]. Human HBV strains occur in nine genotypes A-I, and its major HBV surface antigen (HBsAg) has several immune protective conformational B cell epitopes a, d or y, w1–4 or r [27]. The entire amino acid sequences of each protein from different genotypes were obtained from the UniProt database and aligned in Figure 2.
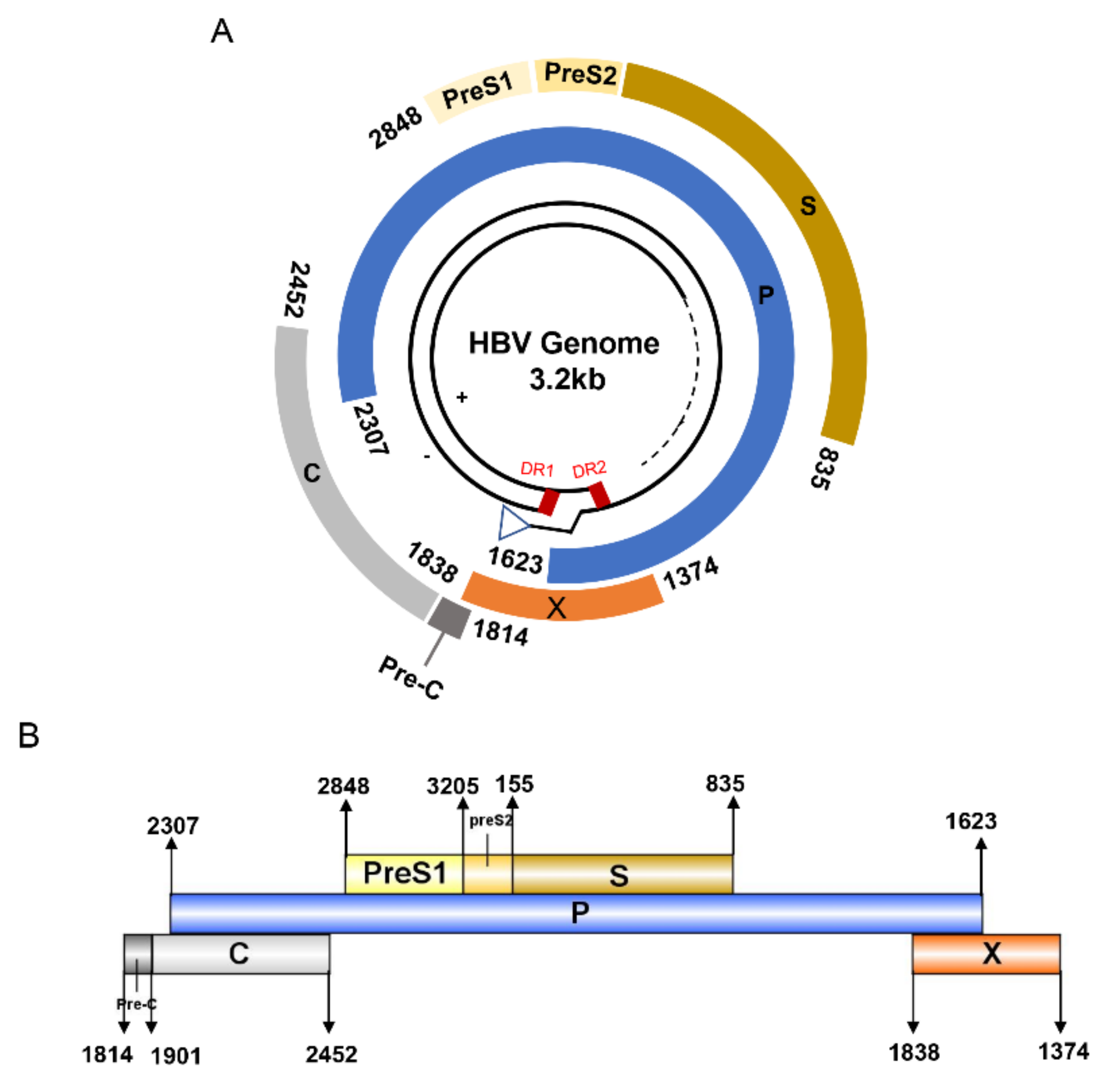
Figure 1. The circular (A) and linear (B) diagram of HBV genome.
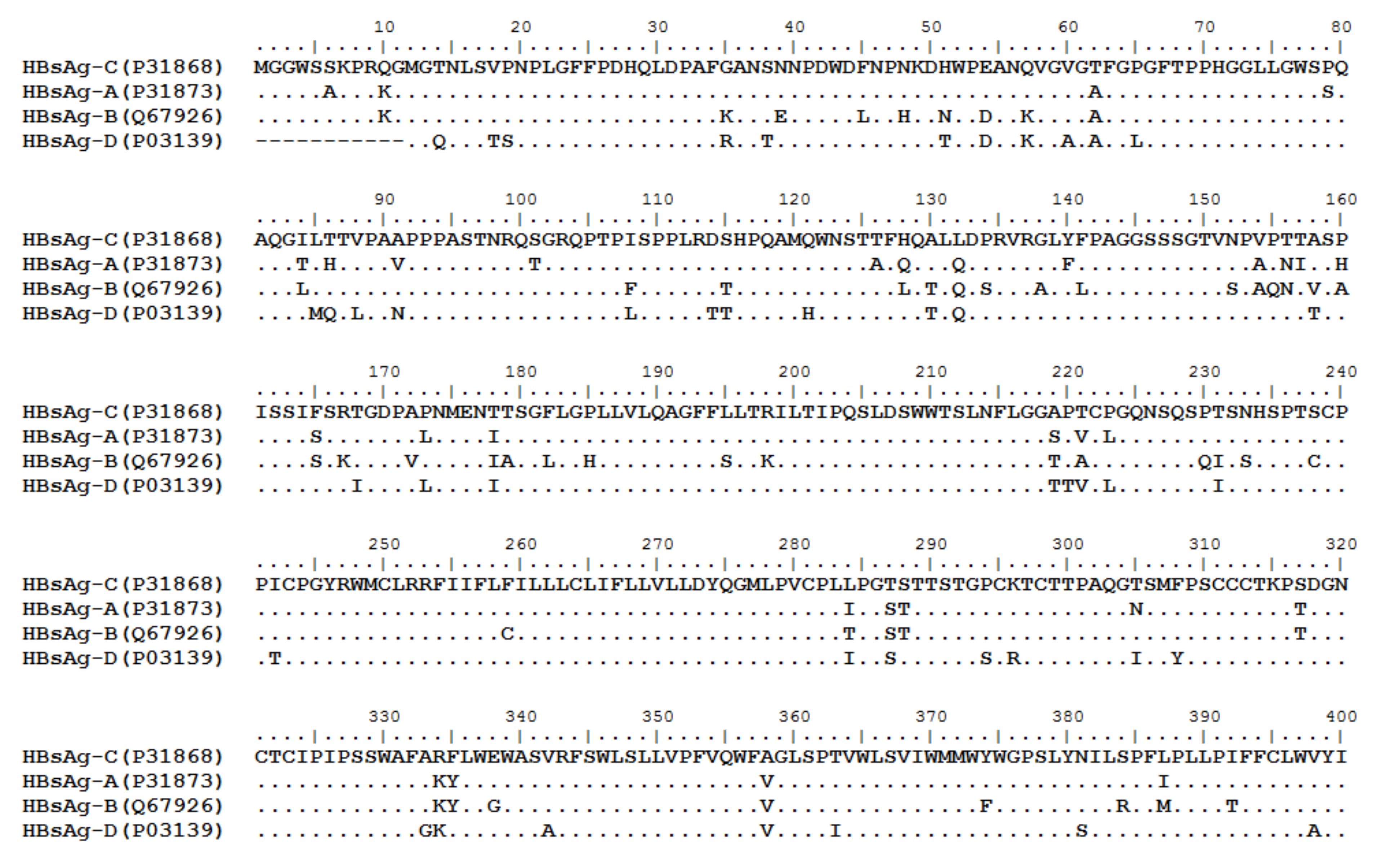
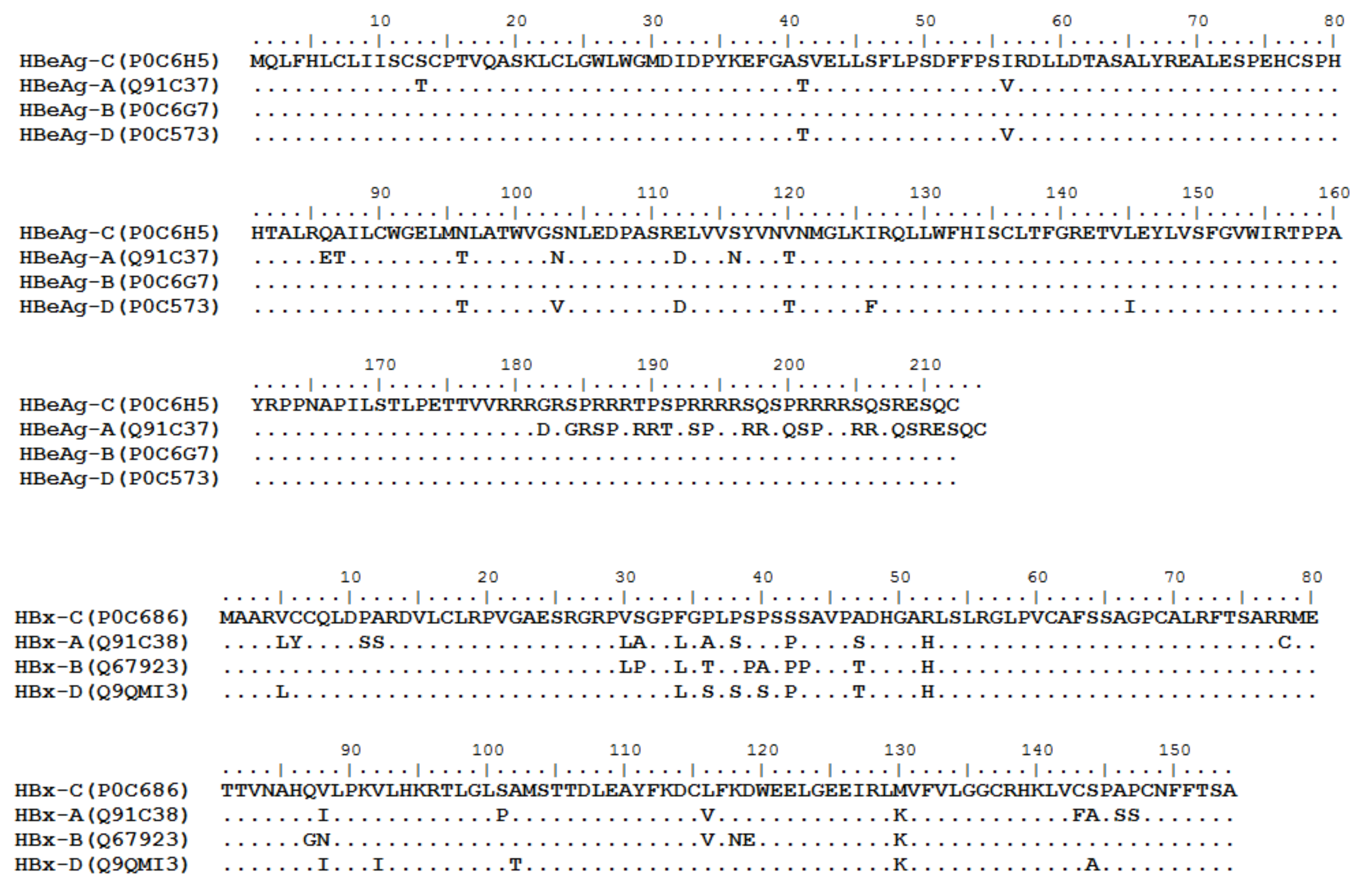
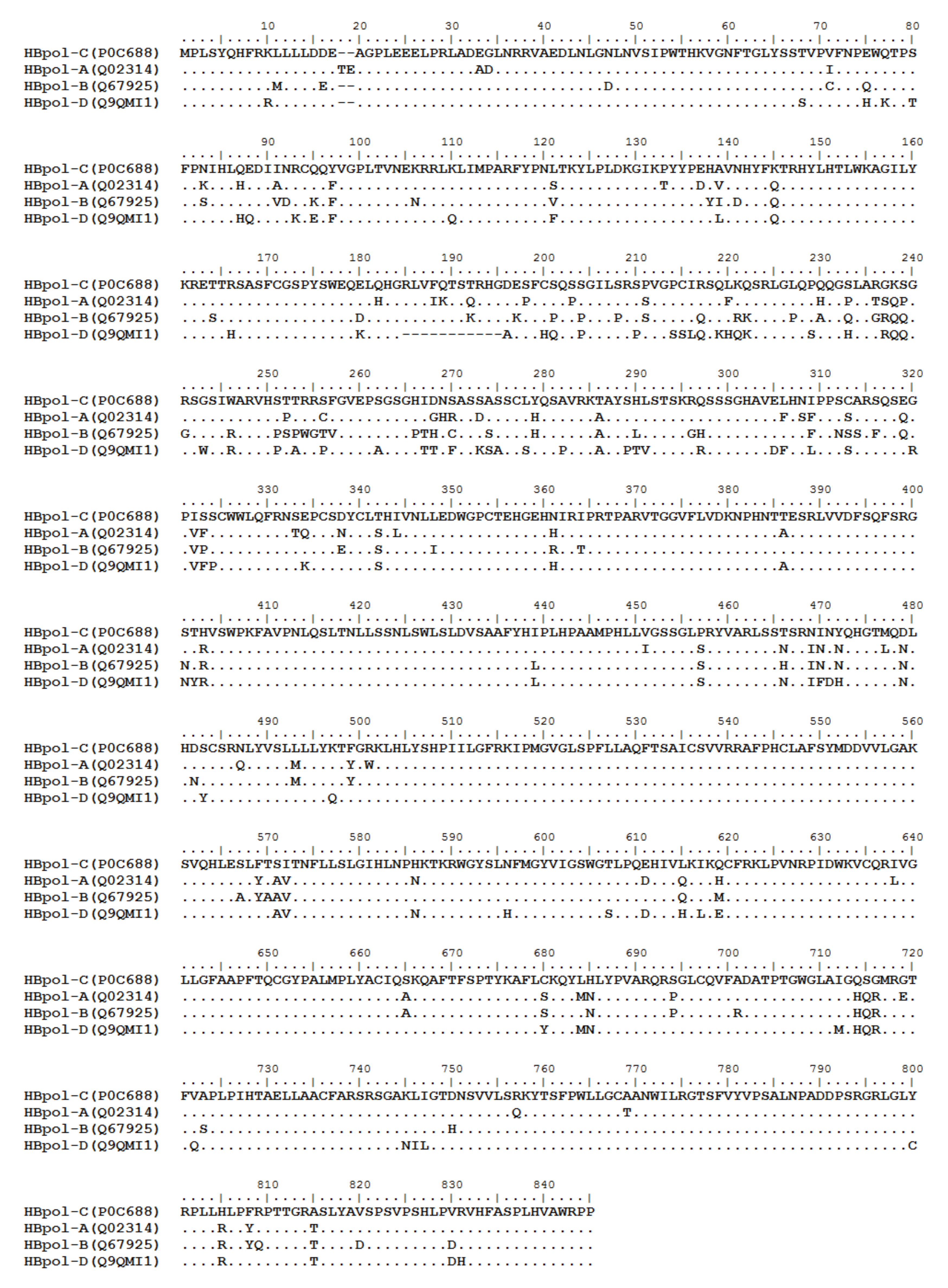
Figure 2. Homologous analysis of HBsAg, HBeAg, HBx and HBpol proteins from HBV C, A, B, and D genotypes. The entire amino acid sequences of each protein from different HBV genotypes were obtained from the UniProt database, aligned and used for in silico prediction of HBV antigen T cell epitopes presented by HLA-A allotypes.
The process of T cell epitope identification begins with the selection of candidate epitope peptides. The first strategy is using overlapping peptides (OLPs) spanning the entire proteome or selected antigens of interest (peptide scanning). Chen et al. expanded HBV-specific T cells in vitro by co-culturing the overlapping peptide pools spanning the entire sequence of HBV genotypes B and C and the peripheral blood mononuclear cells (PBMCs) from patients with chronic HBV infection, followed by the detection of T cell response in each co-culture using IFN-γ enzyme-linked immunospot (IFN-γ ELISpot) assay, IFN-γ intracellular staining and flow cytometry [28]. However, peptide scanning is a high-cost and laborious method due to a large amount of OLPs spanning overall HBV proteins. For CD8 T cell epitopes, HBsAg, HBeAg, HBx and HBpol contain 131, 68, 49, and 279 OLPs, respectively, when overlapping 6 amino acids in each 9-mer peptide. An alternative strategy is to focus on the in silico predicted T cell epitopes binding to the indicated HLA supertypes as calculated by multiple epitope prediction tools and algorithms. Brinck-Jensen et al. predicted 20 HBV-specific epitopes using combined in silico methods and evaluated for the immunogenicity of these epitopes through exposure to patients’ PBMCs by IFN-γ ELISpot [29]. More recently, a similar in silico approach was also employed to assess all previously verified HBx- and HBpol-derived epitopes and to predict novel HLA-binding peptides for 6 HLA supertypes. Then, a part of reported epitopes were chosen for experimental validation. A total of 13 HLA binders derived from HBx and 33 binders from HBpol were described across HLA subtypes by this strategy[30]. Predicted epitopes are based on the indicated HLA restrictions and limit the number of research objects with diverse HLA subtypes to a reasonable range, yet the inaccuracy of theoretical prediction may omit some real-world epitopes.+
The methodologies to validate the immunogenicity of candidate epitope peptides have been improved remarkably over the last two decades. Different assays are utilized for the detection of peptide-induced T cell response or peptide-specific T cells with individual advantages and disadvantages in terms of practicability, cost, sensitivity, function evaluation. The following approaches are currently widely used, such as cytotoxicity assay, proliferation assay, intracellular cytokine staining (ICS), ELISpot/FluoroSpot, and peptide-MHC multimers staining (tetramers, pentamers, or dextramers). The cytotoxicity assay was initially performed to validate CD8 T cell epitopes by co-culturing patients’ PBMCs with target cells labeled with Chromium-51, after the PBMCs were stimulated by the indicated candidate epitope peptides[31]. Additionally, lymphocyte proliferation assay is mostly applied to CD4 T cell epitopes validation. The PBMCs from HBV-infected or HBV-vaccinated individuals were co-cultured with HBV-derived peptides for several days and ++3H-thymidine pulses were administered eventually followed by quantifying the incorporated radioactivity[32]. One more common approach currently utilized is ICS or ELISpot/FluoroSpot. Patient’s PBMCs are in vitro or ex vivo stimulated with the candidate epitope peptides and simultaneously cytokine release is blocked followed by ICS and flow cytometry to define whether CD4 T cells or CD8 T cells activation[33]. The ELISpot or FluoroSpot technology enables the detection of single activated cells among one million PBMCs. The accuracy, sensitivity, reproducibility and durability have led to its widespread applications in researches and the broad prospects in the clinical detection of antigen-specific T cells[34][35] . An issue encountered with ELISpot, FluoroSpot, ICS, and related assays is that they may ignore T cells that produce different cytokines or trace cytokines during the window of time of the assay (e.g., Follicular helper CD4 T cells generally produce very low amounts of cytokines). Peptide-MHC tetramer staining has been the gold standard to quantify antigen-specific T cells with high sensitivity and precision, thus is often used to identify T cell epitopes in many researches. However, the preparation of peptide-MHC tetramers or multimers is high-cost, complicated, and time-consumption[36][37]. A pioneering study focused on all possible peptides of the entire HBV genome and 484 unique HLA-A1101-restricted epitopes predicted by NetMHC algorithms were validated using mass cytometry and multiplex peptide-tetramers staining[38]. Many researchers also have established a transgenic mouse model to map HLA-restricted epitopes. Ru et al. developed and immunized HLA-A2/DP4 mice with epitopes derived from HBsAg to identify four new HLA-DP4-restricted epitopes[39]. Besides cellular functional experiments, peptide-HLA molecule binding and stabilization assays were commonly used to identify epitopes. Pan et al. defined 16 HBV epitopes by analyzing the different binding affinities of candidate epitope peptides with HLA-A3303 using RMA-S cells binding and stability assay. More recently, Ferretti et al. used a high-throughput genome-wide screening technology to identify the target cells expressing candidate epitopes productively recognized by T cells (T-Scan) and determined 29 epitopes in SARS-CoV-2 for the six most prevalent HLA types[40]. Chikata et al. employed immunocapture and liquid chromatography mass spectrometry (LC-MS) subsequent to pre-treatment of the target protein to disrupt its three-dimensional structure to characterize HIV-1 epitope peptides on a large scale presented by HLA-C1202[41]. A variety of epitope assay strategies have been utilized with their own features and potential.+++
References
- Jue Liu; Wannian Liang; Wenzhan Jing; Min Liu; Countdown to 2030: eliminating hepatitis B disease, China. Bulletin of the World Health Organization 2019, 97, 230-238, 10.2471/blt.18.219469.
- Organization, W.H. Global Hepatitis Report 2017. Geneva,switzerland:World Health Organization 2017.
- Claire Pierra Rouviere; Cyril B. Dousson; John E. Tavis; HBV replication inhibitors. Antiviral Research 2020, 179, 104815-104815, 10.1016/j.antiviral.2020.104815.
- Mauro Viganò; Giampaolo Mangia; Pietro Lampertico; HBeAg-negative chronic hepatitis B: why do I treat my patients with nucleos(t)ide analogues?. Liver International 2013, 34, 120-126, 10.1111/liv.12401.
- Maria Buti; HBeAg-positive chronic hepatitis B: Why do I treat my patients with Nucleos(t)ide Analogs?. Liver International 2013, 34, 108-111, 10.1111/liv.12392.
- Cristina Pérez-Cameo; Mònica Pons; Rafael Esteban; New therapeutic perspectives in HBV: when to stop NAs. Liver International 2013, 34, 146-153, 10.1111/liv.12398.
- Masanori Isogawa; Yasuhito Tanaka; Immunobiology of hepatitis B virus infection. Hepatology Research 2014, 45, 179-189, 10.1111/hepr.12439.
- Tai-Chung Tseng; Li-Rung Huang; Immunopathogenesis of Hepatitis B Virus. The Journal of Infectious Diseases 2017, 216, S765-S770, 10.1093/infdis/jix356.
- Yang Cheng; Yuan O. Zhu; Etienne Becht; Pauline Aw; JinMiao Chen; Michael Poidinger; Paola Flórez de Sessions; Martin Lloyd Hibberd; Antonio Bertoletti; Seng Gee Lim; et al.Evan W. Newell Multifactorial heterogeneity of virus-specific T cells and association with the progression of human chronic hepatitis B infection. Science Immunology 2019, 4, eaau6905, 10.1126/sciimmunol.aau6905.
- Xiaochen Wang; QiFeng He; Haiyuan Shen; Xiao-Jie Lu; Beicheng Sun; Genetic and phenotypic difference in CD8+ T cell exhaustion between chronic hepatitis B infection and hepatocellular carcinoma. Journal of Medical Genetics 2018, 56, 18-21, 10.1136/jmedgenet-2018-105267.
- Yuchen Xia; T. Jake Liang; Development of Direct-acting Antiviral and Host-targeting Agents for Treatment of Hepatitis B Virus Infection. Gastroenterology 2019, 156, 311-324, 10.1053/j.gastro.2018.07.057.
- Thjon J. Tang; Jaap Kwekkeboom; Shanta Mancham; Rekha S. Binda; Robert A. de Man; Solko W. Schalm; Johannes G. Kusters; Harry L.A. Janssen; Intrahepatic CD8+ T-lymphocyte response is important for therapy-induced viral clearance in chronic hepatitis B infection. Journal of Hepatology 2005, 43, 45-52, 10.1016/j.jhep.2005.01.038.
- George Papatheodoridis; Ioannis Vlachogiannakos; Evangelos Cholongitas; Karsten Wursthorn; Christos Thomadakis; Giota Touloumi; Jörg Petersen; Discontinuation of oral antivirals in chronic hepatitis B: A systematic review. Hepatology 2016, 63, 1481-1492, 10.1002/hep.28438.
- Pietro Lampertico; Kosh Agarwal; Thomas Berg; Maria Buti; Harry L.A. Janssen; George Papatheodoridis; Fabien Zoulim; Frank Tacke; EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. Journal of Hepatology 2017, 67, 370-398, 10.1016/j.jhep.2017.03.021.
- Laura Rivino; Nina Le Bert; Upkar S. Gill; Kamini Kunasegaran; Yang Cheng; Damien Z.M. Tan; Etienne Becht; Navjyot K. Hansi; Graham R. Foster; Tung-Hung Su; et al.Tai-Chung TsengSeng Gee LimJia-Horng KaoEvan NewellPatrick T.F. KennedyAntonio Bertoletti Hepatitis B virus–specific T cells associate with viral control upon nucleos(t)ide-analogue therapy discontinuation. Journal of Clinical Investigation 2018, 128, 668-681, 10.1172/jci92812.
- Rohit Loomba; T. Jake Liang; Hepatitis B Reactivation Associated With Immune Suppressive and Biological Modifier Therapies: Current Concepts, Management Strategies, and Future Directions. Gastroenterology 2017, 152, 1297-1309, 10.1053/j.gastro.2017.02.009.
- Katja Nitschke; Hendrik Luxenburger; Christoph Neumann-Haefelin; Muthamia M. Kiraithe; Robert Thimme; CD8+ T-Cell Responses in Hepatitis B and C: The (HLA-) A, B, and C of Hepatitis B and C. Digestive Diseases 2016, 34, 396-409, 10.1159/000444555.
- Paul A. Roche; Kazuyuki Furuta; The ins and outs of MHC class II-mediated antigen processing and presentation. Nature Reviews Immunology 2015, 15, 203-216, 10.1038/nri3818.
- Maria Buti; Francisco Rodríguez Frías; Rafael Esteban; Cuantificación del antígeno de superficie del virus de la hepatitis B: implicaciones clínicas. Medicina Clínica 2012, 138, 483-488, 10.1016/j.medcli.2011.04.024.
- Alexandra Alexopoulou; HBeAg negative variants and their role in the natural history of chronic hepatitis B virus infection. World Journal of Gastroenterology 2014, 20, 7644-52, 10.3748/wjg.v20.i24.7644.
- Jing-Hsiung Ou; Molecular biology of hepatitis B virus e antigen. Journal of Gastroenterology and Hepatology 1997, 12, S178-S187, 10.1111/j.1440-1746.1997.tb00499.x.
- L.-Y. Mak; D. K.-H. Wong; Ka-Shing Cheung; W.-K. Seto; Ching-Lung Lai; M.-F. Yuen; Review article: hepatitis B core-related antigen (HBcrAg): an emerging marker for chronic hepatitis B virus infection. Alimentary Pharmacology & Therapeutics 2017, 47, 43-54, 10.1111/apt.14376.
- Ashraf Ali; Hany Abdel-Hafiz; Mohd Suhail; Amany Al-Mars; Mohammad Khalid Zakaria; Kaneez Fatima; Sultan Ahmad; Esam Azhar; Adeel Chaudhary; Ishtiaq Qadri; et al. Hepatitis B virus, HBx mutants and their role in hepatocellular carcinoma. World Journal of Gastroenterology 2014, 20, 10238-48, 10.3748/wjg.v20.i30.10238.
- Yonghe Qi; Zhenchao Gao; Guangwei Xu; Bo Peng; Chenxuan Liu; Huan Yan; Qiyan Yao; Guoliang Sun; Yang Liu; Dingbin Tang; et al.Zilin SongWenhui HeYinyan SunJu-Tao GuoWenhui Li DNA Polymerase κ Is a Key Cellular Factor for the Formation of Covalently Closed Circular DNA of Hepatitis B Virus. PLOS Pathogens 2016, 12, e1005893, 10.1371/journal.ppat.1005893.
- Y Chen; D Yang; S Li; Y Gao; Runqiu Jiang; L Deng; F R Frankel; B Sun; Development of a Listeria monocytogenes-based vaccine against hepatocellular carcinoma. Oncogene 2011, 31, 2140-2152, 10.1038/onc.2011.395.
- Nanna-Sophie Brinck-Jensen; Thomas Vorup-Jensen; Peter Derek Christian Leutscher; Christian Erikstrup; Eskild Petersen; Immunogenicity of twenty peptides representing epitopes of the hepatitis B core and surface antigens by IFN-γ response in chronic and resolved HBV. BMC Immunology 2015, 16, 1-12, 10.1186/s12865-015-0127-7.
- Monique T. A. de Beijer; Diahann T. S. L. Jansen; Yingying Dou; Wim J. E. van Esch; Juk Yee Mok; Mariëlle J. P. Maas; Giso Brasser; Robert A. de Man; Andrea M. Woltman; Sonja I. Buschow; et al. Discovery and Selection of Hepatitis B Virus-Derived T Cell Epitopes for Global Immunotherapy Based on Viral Indispensability, Conservation, and HLA-Binding Strength. Journal of Virology 2020, 94, 1, 10.1128/jvi.01663-19.
- Kimberly Shafer-Weaver; Thomas Sayers; Susan Strobl; Eric Derby; Tracy Ulderich; Michael Baseler; Anatoli Malyguine; The Granzyme B ELISPOT assay: an alternative to the 51Cr-release assay for monitoring cell-mediated cytotoxicity. Journal of Translational Medicine 2003, 1, 14-14, 10.1186/1479-5876-1-14.
- Anthony Pajot; Marie-Louise Michel; Maryline Bourgine; Marie-Noelle Ungeheuer; David Ojcius; Qiang Deng; François A. Lemonnier; Yu-Chun Lone; Identification of novel HLA-DR1-restricted epitopes from the hepatitis B virus envelope protein in mice expressing HLA-DR1 and vaccinated human subjects. Microbes and Infection 2006, 8, 2783-2790, 10.1016/j.micinf.2006.08.009.
- Giulia Freer; Laura Rindi; Intracellular cytokine detection by fluorescence-activated flow cytometry: Basic principles and recent advances. Methods 2013, 61, 30-38, 10.1016/j.ymeth.2013.03.035.
- NianNian Ji; Thomas G. Forsthuber; ELISPOT Techniques. Programmed Necrosis 2014, 1304, 63-71, 10.1007/7651_2014_111.
- Tracy D. Reynolds; Safiehkhatoon Moshkani; Michael D. Robek; An ELISPOT-Based Assay to Measure HBV-Specific CD8+ T Cell Responses in Immunocompetent Mice. Programmed Necrosis 2016, 1540, 237-247, 10.1007/978-1-4939-6700-1_20.
- Garry Dolton; Katie Tungatt; Angharad Lloyd; Valentina Bianchi; Sarah M. Theaker; Andrew Trimby; Christopher J. Holland; Marco Donia; Andrew Godkin; David Cole; et al.Per Thor StratenMark PeakmanInge Marie SvaneAndrew K. Sewell More tricks with tetramers: a practical guide to staining T cells with peptide-MHC multimers. Immunology 2015, 146, 11-22, 10.1111/imm.12499.
- Philip Savage; Maggie Millrain; Sofia Dimakou; Justin Stebbing; Julian Dyson; Expansion of CD8+ Cytotoxic T Cells in vitro and in vivo Using MHC Class I Tetramers. Tumor Biology 2007, 28, 70-76, 10.1159/000099152.
- Antonio Bertoletti; Carlo Ferrari; Adaptive immunity in HBV infection. Journal of Hepatology 2016, 64, S71-S83, 10.1016/j.jhep.2016.01.026.
- Zhitao Ru; Wenjun Xiao; Anthony Pajot; Zhihua Kou; Shihui Sun; Bernard Maillere; Guangyu Zhao; David Ojcius; Yu-Chun Lone; Yusen Zhou; et al. Development of a Humanized HLA-A2.1/DP4 Transgenic Mouse Model and the Use of This Model to Map HLA-DP4-Restricted Epitopes of HBV Envelope Protein. PLoS ONE 2012, 7, e32247, 10.1371/journal.pone.0032247.
- Andrew P. Ferretti; Tomasz Kula; Yifan Wang; Dalena M.V. Nguyen; Adam Weinheimer; Garrett S. Dunlap; Qikai Xu; Nancy Nabilsi; Candace R. Perullo; Alexander W. Cristofaro; et al.Holly J. WhittonAmy VirbasiusKenneth J. OlivierLyndsey R. BucknerAngela T. AlistarEric D. WhitmanSarah A. BertinoShrikantA ChattopadhyayGavin MacBeath Unbiased Screens Show CD8+ T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein. Immunity 2020, 53, 1095-1107.e3, 10.1016/j.immuni.2020.10.006.
- Takayuki Chikata; Wayne Paes; Tomohiro Akahoshi; Thomas Partridge; Hayato Murakoshi; Hiroyuki Gatanaga; Nicola Ternette; Shinichi Oka; Persephone Borrow; Masafumi Takiguchi; et al. Identification of Immunodominant HIV-1 Epitopes Presented by HLA-C*12:02, a Protective Allele, Using an Immunopeptidomics Approach. Journal of Virology 2019, 93, e00634-19, 10.1128/jvi.00634-19.
- Sinu Paul; Nathan P. Croft; Anthony W. Purcell; David C. Tscharke; Alessandro Sette; Morten Nielsen; Bjoern Peters; Benchmarking predictions of MHC class I restricted T cell epitopes in a comprehensively studied model system. PLOS Computational Biology 2020, 16, e1007757, 10.1371/journal.pcbi.1007757.
- Karolina Palucka; Jacques Banchereau; Dendritic-Cell-Based Therapeutic Cancer Vaccines. Immunity 2013, 39, 38-48, 10.1016/j.immuni.2013.07.004.
- Sheila Lumley; Howard Noble; Martin J. Hadley; Liz Callow; Amna Malik; Yi Yi Chua; Owen J. Duffey; Natalia Grolmusova; Arvind Kumar; Samuel Ravenscroft; et al.Jonathan I. SpencerChristoph Neumann-HaefelinRobert ThimmeMonique AnderssonPaul KlenermanEleanor BarnesPhilippa C. Matthews Hepitopes: A live interactive database of HLA class I epitopes in hepatitis B virus. Wellcome Open Research 2016, 1, 1, 10.12688/wellcomeopenres.9952.1.
More
Information
Subjects:
Infectious Diseases
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
3 times
(View History)
Update Date:
19 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





