Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ludovic Bonhomme | + 2243 word(s) | 2243 | 2022-02-11 10:03:11 | | | |
| 2 | Camila Xu | Meta information modification | 2243 | 2022-02-15 03:56:07 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Bonhomme, L. Fusarium graminearum. Encyclopedia. Available online: https://encyclopedia.pub/entry/19425 (accessed on 07 February 2026).
Bonhomme L. Fusarium graminearum. Encyclopedia. Available at: https://encyclopedia.pub/entry/19425. Accessed February 07, 2026.
Bonhomme, Ludovic. "Fusarium graminearum" Encyclopedia, https://encyclopedia.pub/entry/19425 (accessed February 07, 2026).
Bonhomme, L. (2022, February 14). Fusarium graminearum. In Encyclopedia. https://encyclopedia.pub/entry/19425
Bonhomme, Ludovic. "Fusarium graminearum." Encyclopedia. Web. 14 February, 2022.
Copy Citation
Fusarium graminearum, the main causal agent of Fusarium Head Blight (FHB), is one of the most damaging pathogens in wheat. Because of the complex organization of wheat resistance to FHB, this pathosystem represents a relevant model to elucidate the molecular mechanisms underlying plant susceptibility and to identify their main drivers, the pathogen’s effectors.
Fusarium graminearum
Triticum aestivum
plant–fungus interaction
1. Introduction
Fusarium Head Blight (FHB), mainly caused by the Ascomycota fungus Fusarium graminearum, is one of the most prevalent diseases of small grain cereals, especially in wheat [1][2]. With direct impacts on yield, grain quality and through the accumulation of carcinogenic mycotoxins (e.g., deoxynivalenol, DON) [3][4][5], FHB is considered as a major limiting factor for wheat production in Europe, North America and Asia [6][7][8][9], resulting in substantial economic losses that reached up to USD 1.176 billion over 2015 and 2016 in the USA for instance [10]. Because FHB is expected to be even more frequent and intense along with the rises of temperatures and the occasional increases in air humidity promoted through the climate change [11][12], further research is needed to develop better management strategies and sustainable control solutions [13]. FHB resistance trait is strictly quantitative and involves multiple Quantitative Trait Loci (QTLs) with relatively weak effects [14][15] that makes them insufficient when environmental conditions are favorable to the fungus. Thus, identifying sustainable solutions able to efficiently control FHB epidemics requires the search of alternative sources of resistance. For the last twenty years, the multiple evidences of the role of a plant’s susceptibility factors in promoting pathogen infection have opened new opportunities to identify such pivotal determinants of plant diseases, and a number of studies already reported that mutation or loss of susceptibility genes can be used in resistance breeding [16][17]. With the increasing evidences of the role of wheat’s susceptibility factors in FHB development [18][19][20][21][22][23], elucidating the mechanisms of wheat susceptibility to F. graminearum appears as a promising approach to improve FHB resistance [17][24][25][26].
A pathogen’s ability to hijack a host’s biological processes such as defense responses, physiology and primary metabolism to exploit host resources is assumed to be one of the key drivers of a plant’s susceptibility. These interactions involve a complex molecular crosstalk between the two partners, including the delivery of effectors, which include small secreted proteins able to alter host cell structure and to target specific functions into host tissues, the so-called susceptibility factors [27][28][29][30][31][32]. The role of an effector is therefore determined by its in planta localization, i.e., the apoplast or host’s intracellular compartments, and the targeted susceptibility factors [32][33][34][35]. Mining a robust catalog of pathogen effectors, i.e., the effectome, offers major opportunities to improve resistance breeding through the identification of the host’s susceptibility factors, i.e., the targetome. This further could make possible the identification of functional markers to screen plant germplasm, as well as new targets for host-induced gene silencing [32][36][37]. However, their systematic search in silico is still challenging because most of them lack shared protein features or conserved domains within and across species, and very few are structurally characterized [28][32][35]. The only universal fungal effector’s characteristics are their expected secretion and their fine-tuned synthesis along the infection progress [35][38], making in planta exploratory methods such as transcriptomics and proteomics necessary to narrow down the effector candidates and identify the active ones [37][39][40].
Numerous effectors are deployed by pathogens and their role within the molecular crosstalk and in the fate of the interaction is determined by their conservation among pathogen species or between the different strains of a particular species. This conservation is also partly driven by the coevolution with their hosts and their targetome [40][41]. Conserved effectors are thought to play an indispensable role to ensure compatibility by targeting conserved host’s immune or metabolism functions, while specific effectors are thought to be involved in the host’s adaptation and strain aggressiveness [40][41][42][43]. Thus, elucidating the complexity of such molecular crosstalk underlying plant–pathogen interactions and elaborating robust and relevant effectomes require consideration of the diversity from both partners of the interaction. The genomics variability of many fungal pathogen species is well characterized, but its impacts on the infection program remains to be addressed [44][45][46][47][48].
In Fusarium graminearum, genomics variability has been well characterized and the first pangenome of the species, built from 20 strains, was published in 2021 [49]. If the in silico characterization of F. graminearum secretome [49][50] is now available, our knowledge about the effective in planta effectome remains fragmented and needs to be clarified. Several in planta studies outlined a highly dynamic and complex molecular dialogue between wheat and F. graminearum, involving a stage-specific delivery of the effectors [19][25][51][52]. However, the impacts of wheat and F. graminearum genetic backgrounds are largely unknown. In a previous proteomics study, F. graminearum infection strategy was described in three strains of contrasting aggressiveness facing three wheat cultivars of contrasting susceptibility at one time point, resulting in the identification of highly conserved fungal determinants of the infection [20]. Owing to its higher ability in detecting low-abundant molecules, as are the fungal molecules within host tissues, applying RNA-seq technology over infection progress appears as a promising approach to complete the picture of F. graminearum effectome during FHB and to identify its core components.
2. Fusarium graminearum Infection Involves a Highly Conserved Effectome
Transcriptome profiling of effector coding genes conducted on the three F. graminearum strains of contrasting aggressiveness and on the five hosts of contrasting susceptibility to FHB revealed highly conserved effector repertoires. We demonstrated that the three strains shared 90% of their effector-gene transcripts. While at the genomic scale, these three strains shared only 58% of their theoretical secretome [49], our results corroborate a previous in planta proteomics study demonstrating that nearly 100% of the whole identified secreted proteins were accumulated in the same three strains [20]. This emphasizes that the effective infection process of the three strains on wheat is based on a conserved effectome that controls critical plant processes to ensure the success of the infection. Similar results were already found in other pathosystems. For instance, the gene expression analysis of six Puccinia triticina strains highlighted a highly conserved infection strategy with 85.7% of the identified secretome genes expressed by all the strains during wheat infection [44]. Similar findings were also reported in the maize–Exserohilum turcicum interaction where 97% of the putative effector genes were shared by two strains [48]. Extending this analysis to the role of host genetic background on the expressed effectome, our data also demonstrated a highly conserved infection strategy in different wheat cultivars of contrasting susceptibility to FHB that engages a common effector repertoire shared at 91%. Few hosts’ specific gene expressions were already outlined at the whole-transcriptome scale [53]. Our results support similar conclusions with a special focus on the effectome gene set and are consistent with our previous proteomics study demonstrating the accumulation of the same fungal proteins in different wheat hosts [20]. A core effectome composed of 357 genes expressed by all the strains and in all wheat hosts (Figure 1) exemplified the highly conserved infection program established by F. graminearum. Because the interaction is systematically producing FHB disease regardless of the strain aggressiveness or the host susceptibility, these genes likely include key drivers of FHB in bread wheat. Their functions are thus supposed to be crucial determinants of basal processes powering the FHB development in wheat, including 66 putative effector genes and 21 Phi-base matches known to be involved in pathogenicity.
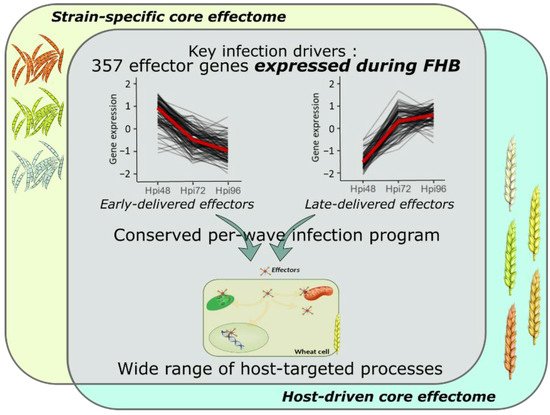
Figure 1. Model summarizing the conserved and complex F. graminearum infection strategy on wheat spikes. As a whole, 357 effector genes were identified as the key drivers of FHB infection expressed by all the strains and in all the infected hosts; they represent the F. graminearum core effectome. These genes were expressed at very specific infection stages in a per-wave manner, including genes highly expressed at the very beginning of the interaction with the wheat tissues and others highly expressed in the later stages of the infection. The timing of gene expression was mostly conserved independently of the strain or the host. Targeted processes within the host are highly diverse with a wide array of targeted compartments and predicted functions.
3. F. graminearum Core Effectors Are Delivered in a Conservative Per-Wave Expression
The core-effectome demonstrated to be deeply remodeled along with the infection progress, depicting the dynamic nature of its components. As previously shown, putative effector proteins were proved to be accumulated at specific stages of the infection process evidencing a specific transition that distinguishes early from late protein accumulations [19]. In line with this previous work, we also observed changing gene expression patterns at the same time (48 to 72 hpi transition), thus corroborating the major reorganization of the molecular arsenal that drives FHB infection. Furthermore, the fine-tuned timing of gene expression was mostly preserved in terms of dynamics for all the strains independently of their aggressiveness and in all the infected hosts independently of their susceptibility level, suggesting that F. graminearum set up a widely conserved genetic program with crucial functions required at very precise infection stages (Figure 1). This conserved infection program may be representative of F. graminearum generalist lifestyle, i.e., interacting with a wide range of hosts and spreading in different tissues [2][54], which results in a lower selection pressure and coevolution with a specific host species [43][55][56].
Besides these conserved infection patterns, some specific regulations in effector-genes were also found in the different fungal strains, but no clear link between gene expression magnitude and aggressiveness has been observed. Identified effector-genes were mainly located in the fast-evolving part of F. graminearum genome, characterized by genes of shorter size, larger variations in exon content and a higher proportion of synonymous and nonsynonymous mutations, together with genes known to be highly transcribed during plant infection in comparison with fungal vegetative growth [49][57][58]. In our study, chromosome 2 for instance, displaying the highest density of polymorphism, also exhibited the highest effector gene density [57]. This polymorphism could explain a part of the observed strain-specific effects on effector-genes expression levels and further protein accumulations. Moreover, intrinsic characteristics of both MDC_Fg1, i.e., a French isolate [59], and ‘Chinese Spring’, i.e., an Asian spring cultivar, might be the main cause of F. graminearum specific expression patterns of the late-delivered effectors observed when facing ‘Chinese Spring’ in comparison to the European winter wheat cultivars, suggesting a remarkable ability to adapt to the different molecular contexts expressed in different wheat cultivars.
4. F. graminearum Infection Strategy Involves Integrative Host Cellular Processes
The search for localization signals within the F. graminearum secreted protein sequences revealed that putative effectors can target host apoplast, as well as different subcellular compartments including nucleus, chloroplast and mitochondria at several infection stages (Figure 7). Along with the relatively high diversity of predicted functions (66 GO terms and 140 Pfam), this supports that the infection success is based on a wide array of manipulated host pathways and echoes previous studies that evidenced the diverse nature of processes involved in FHB susceptibility [19][20][23].
Host apoplast appeared as the main target of the F. graminearum core effectome, including 53 putative genes with additional effector features, i.e., small cysteine-rich proteins. These genes gathered 65 CAZymes that depict the role of cell-wall degradation during FHB to promote host colonization and nutrient acquisition [60]. Eighteen others belonged to peptidases suggesting that F. graminearum is able to override host defense mechanisms especially by interacting with chitin and glucan-triggered immunity and inhibiting host enzymes and proteases as well as to acquire nutrients [61][62][63][64]. Besides these proteases, a guanine-specific ribonuclease was also predicted as a core apoplastic putative effector extending the control of plant stress responses to secreted nucleotidases [60][65]. In addition, three killer toxin KP4-like genes were also identified. Although a previous work has already shown their upregulation during wheat seedling rot disease and FHB, their role in virulence was proved only in seedling rot disease [66].
Intracellular core effectors of F. graminearum mainly targeted host nucleus, including two that match with validated virulence factors, a cysteine-rich secretory protein [67] and a PhoD-like phosphatase protein [68][69] along with one gene with additional effector features, i.e., a small cysteine rich protein. Through its eight predicted core nuclear proteases, F. graminearum might reprogram host gene expression by interfering with the plant’s transcription factors. This strategy was already found in the pathogenic bacteria Xanthomonas euvesicatoria and Pseudomonas syringae that target transcription factors involved in phytohormone pathways [70][71]. Nuclear effectors are also known to act on host transcription machinery by a direct binding on DNA, such as the Melampsora larici-populina Mlp124478 effector that represses genes involved in defense mechanisms [72]. A same strategy might be involved in the F. graminearum infection process though its own nuclear effectors.
Chloroplast and mitochondria were also important targets of F. graminearum core effectome, including three and one genes encoding small cysteine rich proteins, respectively, as well as a putative mitochondrial PhoD-like phosphatase virulence factor [68][69]. These organelles represent important biological hubs interconnecting primary metabolism, energy production, signaling pathways and plant responses to stress [73][74]. The inhibition of the defense mechanisms through the manipulation of chloroplast [75] and mitochondrial [76] processes was already evidenced in several plant–fungi interactions and proved here to be part of the F. graminearum infection strategy. In the case of the chloroplast, its central role has already been described in previous FHB studies [19][20][23]. Finally, effectors with multiple host targets were also detected, suggesting that one effector can achieve completely different functions during the infection progress. An effector targeting both the chloroplast and the mitochondria was validated in poplar—Melampsora larici-populina [77][78] and, as it was outlined in Blumeria graminis f. sp. hordei with the BEC1054 RNase-like effector, those versatile effectors seem to disturb one specific process, such as a host’s defense mechanisms, at several levels by interacting with multiple host proteins [79].
References
- Parry, D.W.; Jenkinson, P.; McLeod, L. Fusarium Ear Blight (Scab) in Small Grain Cereals—A Review. Plant Pathol. 1995, 44, 207–238.
- Goswami, R.S.; Kistler, H.C. Heading for Disaster: Fusarium Graminearum on Cereal Crops. Mol. Plant Pathol. 2004, 5, 515–525.
- Boyacioǧlu, D.; Hettiarachchy, N.S. Changes in Some Biochemical Components of Wheat Grain That Was Infected with Fusarium Graminearum. J. Cereal Sci. 1995, 21, 57–62.
- Argyris, J.; Sanford, D.; TeKrony, D. Fusarium Graminearum Infection during Wheat Seed Development and Its Effect on Seed Quality. Crop Sci. 2003, 43, 1782–1788.
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium Graminearum Trichothecene Mycotoxins: Biosynthesis, Regulation, and Management. Annu. Rev. Phytopathol. 2019, 57, 15–39.
- Dill-Macky, R. Chapter1—Fusarium Head Blight: Recent Epidemics and Research Efforts in the Upper Midwest of the United States. In Fusarium Head Scab: Global Status and Future Prospects; Dubin, H.J., Gilchrist, L., Reeves, J., McNab, A., Eds.; CIMMYT: El Batán, Mexico, 1997.
- Windels, C.E. Economic and Social Impacts of Fusarium Head Blight: Changing Farms and Rural Communities in the Northern Great Plains. Phytopathology 2000, 90, 17–21.
- McMullen, M.; Bergstrom, G.; De Wolf, E.; Dill-Macky, R.; Hershman, D.; Shaner, G.; Van Sanford, D. A Unified Effort to Fight an Enemy of Wheat and Barley: Fusarium Head Blight. Plant Dis. 2012, 96, 1712–1728.
- Dahl, B.; Wilson, W.W. Risk Premiums Due to Fusarium Head Blight (FHB) in Wheat and Barley. Agric. Syst. 2018, 162, 145–153.
- Wilson, W.; Dahl, B.; Nganje, W. Economic Costs of Fusarium Head Blight, Scab and Deoxynivalenol. World Mycotoxin J. 2018, 11, 291–302.
- Vaughan, M.; Backhouse, D.; Ponte, E.M.D. Climate Change Impacts on the Ecology of Fusarium Graminearum Species Complex and Susceptibility of Wheat to Fusarium Head Blight: A Review. World Mycotoxin J. 2016, 9, 685–700.
- Mylonas, I.; Stavrakoudis, D.; Katsantonis, D.; Korpetis, E. Chapter 1—Better farming practices to combat climate change. In Climate Change and Food Security with Emphasis on Wheat, 1st ed.; Ozturk, M., Gul, A., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 1–29.
- Xia, R.; Schaafsma, A.W.; Wu, F.; Hooker, D.C. Impact of the Improvements in Fusarium Head Blight and Agronomic Management on Economics of Winter Wheat. World Mycotoxin J. 2020, 13, 423–439.
- Venske, E.; dos Santos, R.S.; Farias, D.d.R.; Rother, V.; da Maia, L.C.; Pegoraro, C.; Costa de Oliveira, A. Meta-Analysis of the QTLome of Fusarium Head Blight Resistance in Bread Wheat: Refining the Current Puzzle. Front. Plant Sci. 2019, 10, 727.
- Zheng, T.; Hua, C.; Li, L.; Sun, Z.; Yuan, M.; Bai, G.; Humphreys, G.; Li, T. Integration of Meta-QTL Discovery with Omics: Towards a Molecular Breeding Platform for Improving Wheat Resistance to Fusarium Head Blight. Crop J. 2021, 9, 739–749.
- Pavan, S.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Loss of Susceptibility as a Novel Breeding Strategy for Durable and Broad-Spectrum Resistance. Mol. Breed. 2010, 25, 1–12.
- van Schie, C.C.N.; Takken, F.L.W. Susceptibility Genes 101: How to Be a Good Host. Annu. Rev. Phytopathol. 2014, 52, 551–581.
- Ma, H.-X.; Bai, G.-H.; Gill, B.S.; Hart, L.P. Deletion of a Chromosome Arm Altered Wheat Resistance to Fusarium Head Blight and Deoxynivalenol Accumulation in Chinese Spring. Plant Dis. 2006, 90, 1545–1549.
- Fabre, F.; Vignassa, M.; Urbach, S.; Langin, T.; Bonhomme, L. Time-resolved Dissection of the Molecular Crosstalk Driving Fusarium Head Blight in Wheat Provides New Insights into Host Susceptibility Determinism. Plant Cell Environ. 2019, 42, 2291–2308.
- Fabre, F.; Bormann, J.; Urbach, S.; Roche, S.; Langin, T.; Bonhomme, L. Unbalanced Roles of Fungal Aggressiveness and Host Cultivars in the Establishment of the Fusarium Head Blight in Bread Wheat. Front. Microbiol. 2019, 10, 2857.
- Su, Z.; Bernardo, A.; Tian, B.; Chen, H.; Wang, S.; Ma, H.; Cai, S.; Liu, D.; Zhang, D.; Li, T.; et al. A Deletion Mutation in TaHRC Confers Fhb1 Resistance to Fusarium Head Blight in Wheat. Nat. Genet. 2019, 51, 1099–1105.
- Hales, B.; Steed, A.; Giovannelli, V.; Burt, C.; Lemmens, M.; Molnár-Láng, M.; Nicholson, P. Type II Fusarium Head Blight Susceptibility Conferred by a Region on Wheat Chromosome 4D. J. Exp. Bot. 2020, 71, 4703–4714.
- Fabre, F.; Urbach, S.; Roche, S.; Langin, T.; Bonhomme, L. Proteomics-Based Data Integration of Wheat Cultivars Facing Fusarium Graminearum Strains Revealed a Core-Responsive Pattern Controlling Fusarium Head Blight. Front. Plant Sci. 2021, 12, 644810.
- Zaidi, S.S.-A.; Mukhtar, M.S.; Mansoor, S. Genome Editing: Targeting Susceptibility Genes for Plant Disease Resistance. Trends Biotechnol. 2018, 36, 898–906.
- Fabre, F.; Rocher, F.; Alouane, T.; Langin, T.; Bonhomme, L. Searching for FHB Resistances in Bread Wheat: Susceptibility at the Crossroad. Front. Plant Sci. 2020, 11, 731.
- Gorash, A.; Armonienė, R.; Kazan, K. Can Effectoromics and Loss-of-Susceptibility Be Exploited for Improving Fusarium Head Blight Resistance in Wheat? Crop J. 2021, 9, 1–16.
- O’Connell, R.J.; Panstruga, R. Tête à Tête inside a Plant Cell: Establishing Compatibility between Plants and Biotrophic Fungi and Oomycetes. New Phytol. 2006, 171, 699–718.
- Selin, C.; de Kievit, T.R.; Belmonte, M.F.; Fernando, W.G.D. Elucidating the Role of Effectors in Plant-Fungal Interactions: Progress and Challenges. Front. Microbiol. 2016, 7, 600.
- Zeilinger, S.; Gupta, V.K.; Dahms, T.E.S.; Silva, R.N.; Singh, H.B.; Upadhyay, R.S.; Gomes, E.V.; Tsui, C.K.-M.; Nayak, S.C. Friends or Foes? Emerging Insights from Fungal Interactions with Plants. FEMS Microbiol. Rev. 2016, 40, 182–207.
- Khan, M.; Seto, D.; Subramaniam, R.; Desveaux, D. Oh, the Places They’ll Go! A Survey of Phytopathogen Effectors and Their Host Targets. Plant J. 2018, 93, 651–663.
- He, Q.; McLellan, H.; Boevink, P.C.; Birch, P.R.J. All Roads Lead to Susceptibility: The Many Modes of Action of Fungal and Oomycete Intracellular Effectors. Plant Commun. 2020, 1, 100050.
- Jaswal, R.; Kiran, K.; Rajarammohan, S.; Dubey, H.; Singh, P.K.; Sharma, Y.; Deshmukh, R.; Sonah, H.; Gupta, N.; Sharma, T.R. Effector Biology of Biotrophic Plant Fungal Pathogens: Current Advances and Future Prospects. Microbiol. Res. 2020, 241, 126567.
- Brown, N.A.; Hammond-Kosack, K.E. Chapter 19—Secreted biomolecules in fungal plant pathogenesis. In Fungal Biomolecules: Sources, Applications and Recent Developments; Gupta, V.K., Mach, R.L., Sreenivasaprasad, S., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 263–310.
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal Effectors and Plant Susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545.
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Manners, J.M.; Singh, K.B.; Taylor, J.M. Advances and Challenges in Computational Prediction of Effectors from Plant Pathogenic Fungi. PLoS Pathog. 2015, 11, e1004806.
- Nejat, N.; Rookes, J.; Mantri, N.L.; Cahill, D.M. Plant–Pathogen Interactions: Toward Development of next-Generation Disease-Resistant Plants. Crit. Rev. Biotechnol. 2017, 37, 229–237.
- Prasad, P.; Savadi, S.; Bhardwaj, S.C.; Gangwar, O.P.; Kumar, S. Rust Pathogen Effectors: Perspectives in Resistance Breeding. Planta 2019, 250, 1–22.
- Uhse, S.; Djamei, A. Effectors of Plant-Colonizing Fungi and Beyond. PLoS Pathog. 2018, 14, e1006992.
- Lorrain, C.; Gonçalves dos Santos, K.C.; Germain, H.; Hecker, A.; Duplessis, S. Advances in Understanding Obligate Biotrophy in Rust Fungi. New Phytol. 2019, 222, 1190–1206.
- Depotter, J.R.L.; Doehlemann, G. Target the Core: Durable Plant Resistance against Filamentous Plant Pathogens through Effector Recognition. Pest Manag. Sci. 2020, 76, 426–431.
- Arroyo-Velez, N.; González-Fuente, M.; Peeters, N.; Lauber, E.; Noël, L.D. From Effectors to Effectomes: Are Functional Studies of Individual Effectors Enough to Decipher Plant Pathogen Infectious Strategies? PLoS Pathog. 2020, 16, e1009059.
- Badet, T.; Croll, D. The Rise and Fall of Genes: Origins and Functions of Plant Pathogen Pangenomes. Curr. Opin. Plant Biol. 2020, 56, 65–73.
- Petre, B.; Lorrain, C.; Stukenbrock, E.H.; Duplessis, S. Host-Specialized Transcriptome of Plant-Associated Organisms. Curr. Opin. Plant Biol. 2020, 56, 81–88.
- Bruce, M.; Neugebauer, K.A.; Joly, D.L.; Migeon, P.; Cuomo, C.A.; Wang, S.; Akhunov, E.; Bakkeren, G.; Kolmer, J.A.; Fellers, J.P. Using Transcription of Six Puccinia Triticina Races to Identify the Effective Secretome during Infection of Wheat. Front. Plant Sci. 2014, 4, 520.
- Kamel, L.; Tang, N.; Malbreil, M.; San Clemente, H.; Le Marquer, M.; Roux, C.; Frei dit Frey, N. The Comparison of Expressed Candidate Secreted Proteins from Two Arbuscular Mycorrhizal Fungi Unravels Common and Specific Molecular Tools to Invade Different Host Plants. Front. Plant Sci. 2017, 8, 124.
- Rutter, W.B.; Salcedo, A.; Akhunova, A.; He, F.; Wang, S.; Liang, H.; Bowden, R.L.; Akhunov, E. Divergent and Convergent Modes of Interaction between Wheat and Puccinia Graminis f. Sp. Tritici Isolates Revealed by the Comparative Gene Co-Expression Network and Genome Analyses. BMC Genom. 2017, 18, 291.
- Haueisen, J.; Möller, M.; Eschenbrenner, C.J.; Grandaubert, J.; Seybold, H.; Adamiak, H.; Stukenbrock, E.H. Highly Flexible Infection Programs in a Specialized Wheat Pathogen. Ecol. Evol. 2019, 9, 275–294.
- Human, M.P.; Berger, D.K.; Crampton, B.G. Time-Course RNAseq Reveals Exserohilum Turcicum Effectors and Pathogenicity Determinants. Front. Microbiol. 2020, 11, 360.
- Alouane, T.; Rimbert, H.; Bormann, J.; González-Montiel, G.A.; Loesgen, S.; Schäfer, W.; Freitag, M.; Langin, T.; Bonhomme, L. Comparative Genomics of Eight Fusarium graminearum Strains with Contrasting Aggressiveness Reveals an Expanded Open Pangenome and Extended Effector Content Signatures. Int. J. Mol. Sci. 2021, 22, 6257.
- Brown, N.A.; Antoniw, J.; Hammond-Kosack, K.E. The Predicted Secretome of the Plant Pathogenic Fungus Fusarium graminearum: A Refined Comparative Analysis. PLoS ONE 2012, 7, e33731.
- Lysøe, E.; Seong, K.-Y.; Kistler, H.C. The Transcriptome of Fusarium graminearum During the Infection of Wheat. Mol. Plant Microbe Interact. 2011, 24, 995–1000.
- Brown, N.A.; Evans, J.; Mead, A.; Hammond-Kosack, K.E. A Spatial Temporal Analysis of the Fusarium graminearum Transcriptome during Symptomless and Symptomatic Wheat Infection: Transcriptome of Symptomless Fusarium Infection. Mol. Plant Pathol. 2017, 18, 1295–1312.
- Pan, Y.; Liu, Z.; Rocheleau, H.; Fauteux, F.; Wang, Y.; McCartney, C.; Ouellet, T. Transcriptome Dynamics Associated with Resistance and Susceptibility against Fusarium Head Blight in Four Wheat Genotypes. BMC Genom. 2018, 19, 642.
- Li, H.B.; Xie, G.Q.; Ma, J.; Liu, G.R.; Wen, S.M.; Ban, T.; Chakraborty, S.; Liu, C.J. Genetic Relationships between Resistances to Fusarium Head Blight and Crown Rot in Bread Wheat (Triticum Aestivum L.). Theor. Appl. Genet. 2010, 121, 941–950.
- Woolhouse, M.E.J.; Webster, J.P.; Domingo, E.; Charlesworth, B.; Levin, B.R. Biological and Biomedical Implications of the Co-Evolution of Pathogens and Their Hosts. Nat. Genet. 2002, 32, 569–577.
- Newman, T.E.; Derbyshire, M.C. The Evolutionary and Molecular Features of Broad Host-Range Necrotrophy in Plant Pathogenic Fungi. Front. Plant Sci. 2020, 11, 591733.
- Laurent, B.; Moinard, M.; Spataro, C.; Ponts, N.; Barreau, C.; Foulongne-Oriol, M. Landscape of Genomic Diversity and Host Adaptation in Fusarium graminearum. BMC Genom. 2017, 18, 203.
- Wang, Q.; Jiang, C.; Wang, C.; Chen, C.; Xu, J.-R.; Liu, H. Characterization of the Two-Speed Subgenomes of Fusarium graminearum Reveals the Fast-Speed Subgenome Specialized for Adaption and Infection. Front. Plant Sci. 2017, 8, 140.
- Alouane, T.; Rimbert, H.; Fabre, F.; Cambon, F.; Langin, T.; Bonhomme, L. Genome Sequence of Fusarium graminearum Strain MDC_Fg1, Isolated from Bread Wheat Grown in France. Microbiol. Resour. Announc. 2018, 7, e01260-18.
- Wang, Y.; Wang, Y. Trick or Treat: Microbial Pathogens Evolved Apoplastic Effectors Modulating Plant Susceptibility to Infection. Mol. Plant Microbe Interact. 2017, 31, 6–12.
- Jashni, M.K.; Dols, I.H.M.; Iida, Y.; Boeren, S.; Beenen, H.G.; Mehrabi, R.; Collemare, J.; de Wit, P.J.G.M. Synergistic Action of a Metalloprotease and a Serine Protease from Fusarium oxysporum f. sp. lycopersici Cleaves Chitin-Binding Tomato Chitinases, Reduces Their Antifungal Activity, and Enhances Fungal Virulence. Mol. Plant Microbe Interact. 2015, 28, 996–1008.
- Jashni, M.K.; Mehrabi, R.; Collemare, J.; Mesarich, C.H.; de Wit, P.J.G.M. The Battle in the Apoplast: Further Insights into the Roles of Proteases and Their Inhibitors in Plant–Pathogen Interactions. Front. Plant Sci. 2015, 6, 584.
- Muszewska, A.; Stepniewska-Dziubinska, M.M.; Steczkiewicz, K.; Pawlowska, J.; Dziedzic, A.; Ginalski, K. Fungal Lifestyle Reflected in Serine Protease Repertoire. Sci. Rep. 2017, 7, 9147.
- Jashni, M.K.; van der Burgt, A.; Battaglia, E.; Mehrabi, R.; Collemare, J.; de Wit, P.J.G.M. Transcriptome and Proteome Analyses of Proteases in Biotroph Fungal Pathogen Cladosporium fulvum. J. Plant Pathol. 2020, 102, 377–386.
- Rocafort, M.; Fudal, I.; Mesarich, C.H. Apoplastic Effector Proteins of Plant-Associated Fungi and Oomycetes. Curr. Opin. Plant Biol. 2020, 56, 9–19.
- Lu, S.; Faris, J.D. Fusarium graminearum KP4-like Proteins Possess Root Growth-Inhibiting Activity against Wheat and Potentially Contribute to Fungal Virulence in Seedling Rot. Fungal Genet. Biol. 2019, 123, 1–13.
- Lu, S.; Edwards, M.C. Molecular Characterization and Functional Analysis of PR-1-Like Proteins Identified from the Wheat Head Blight Fungus Fusarium graminearum. Phytopathology 2018, 108, 510–520.
- Yun, Y.; Liu, Z.; Yin, Y.; Jiang, J.; Chen, Y.; Xu, J.-R.; Ma, Z. Functional Analysis of the Fusarium graminearum Phosphatome. New Phytol. 2015, 207, 119–134.
- Zhang, Y.; He, J.; Jia, L.-J.; Yuan, T.-L.; Zhang, D.; Guo, Y.; Wang, Y.; Tang, W.-H. Cellular Tracking and Gene Profiling of Fusarium graminearum during Maize Stalk Rot Disease Development Elucidates Its Strategies in Confronting Phosphorus Limitation in the Host Apoplast. PLoS Pathog. 2016, 12, e1005485.
- Kim, J.-G.; Stork, W.; Mudgett, M.B. Xanthomonas Type III Effector XopD Desumoylates Tomato Transcription Factor SlERF4 to Suppress Ethylene Responses and Promote Pathogen Growth. Cell Host Microbe 2013, 13, 143–154.
- Gimenez-Ibanez, S.; Boter, M.; Fernández-Barbero, G.; Chini, A.; Rathjen, J.P.; Solano, R. The Bacterial Effector HopX1 Targets JAZ Transcriptional Repressors to Activate Jasmonate Signaling and Promote Infection in Arabidopsis. PLoS Biol. 2014, 12, e1001792.
- Ahmed, M.B.; dos Santos, K.C.G.; Sanchez, I.B.; Petre, B.; Lorrain, C.; Plourde, M.B.; Duplessis, S.; Desgagné-Penix, I.; Germain, H. A Rust Fungal Effector Binds Plant DNA and Modulates Transcription. Sci. Rep. 2018, 8, 14718.
- Lu, Y.; Yao, J. Chloroplasts at the Crossroad of Photosynthesis, Pathogen Infection and Plant Defense. Int. J. Mol. Sci. 2018, 19, 3900.
- Van Aken, O. Mitochondrial Redox Systems as Central Hubs in Plant Metabolism and Signaling. Plant Physiol. 2021, 186, 36–52.
- Xu, Q.; Tang, C.; Wang, X.; Sun, S.; Zhao, J.; Kang, Z.; Wang, X. An Effector Protein of the Wheat Stripe Rust Fungus Targets Chloroplasts and Suppresses Chloroplast Function. Nat. Commun. 2019, 10, 5571.
- Xu, G.; Zhong, X.; Shi, Y.; Liu, Z.; Jiang, N.; Liu, J.; Ding, B.; Li, Z.; Kang, H.; Ning, Y.; et al. A Fungal Effector Targets a Heat Shock–Dynamin Protein Complex to Modulate Mitochondrial Dynamics and Reduce Plant Immunity. Sci. Adv. 2020, 6, eabb7719.
- Petre, B.; Saunders, D.G.O.; Sklenar, J.; Lorrain, C.; Win, J.; Duplessis, S.; Kamoun, S. Candidate Effector Proteins of the Rust Pathogen Melampsora larici-populina Target Diverse Plant Cell Compartments. Mol. Plant Microbe Interact. 2015, 28, 689–700.
- Petre, B.; Lorrain, C.; Saunders, D.G.O.; Win, J.; Sklenar, J.; Duplessis, S.; Kamoun, S. Rust Fungal Effectors Mimic Host Transit Peptides to Translocate into Chloroplasts. Cell. Microbiol. 2016, 18, 453–465.
- Pennington, H.G.; Gheorghe, D.M.; Damerum, A.; Pliego, C.; Spanu, P.D.; Cramer, R.; Bindschedler, L.V. Interactions between the Powdery Mildew Effector BEC1054 and Barley Proteins Identify Candidate Host Targets. J. Proteome Res. 2016, 15, 826–839.
More
Information
Subjects:
Microbiology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.4K
Revisions:
2 times
(View History)
Update Date:
15 Feb 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




