Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Zhaohui Xue | + 2823 word(s) | 2823 | 2022-02-07 09:46:55 | | | |
| 2 | Rita Xu | Meta information modification | 2823 | 2022-02-15 03:42:45 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Xue, Z. Auxin Response Factors. Encyclopedia. Available online: https://encyclopedia.pub/entry/19416 (accessed on 08 February 2026).
Xue Z. Auxin Response Factors. Encyclopedia. Available at: https://encyclopedia.pub/entry/19416. Accessed February 08, 2026.
Xue, Zhaohui. "Auxin Response Factors" Encyclopedia, https://encyclopedia.pub/entry/19416 (accessed February 08, 2026).
Xue, Z. (2022, February 14). Auxin Response Factors. In Encyclopedia. https://encyclopedia.pub/entry/19416
Xue, Zhaohui. "Auxin Response Factors." Encyclopedia. Web. 14 February, 2022.
Copy Citation
Auxin response factors (ARFs) are an important family of transcription factors in the auxin signaling pathway that are involved in the exertion of auxin in plants and play a key role in regulating plant growth and development.
IAA
auxin
plant hormone
transcription factor
1. Introduction
Auxin is the first discovered plant hormone, one that regulates plant growth and development extensively. The function of auxin depends on a signal transduction pathway involving a variety of transcription factors, one of the most important of which is auxin response factor (ARF). Since the ARF was discovered in Arabidopsis in 1997, ARF has been found in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), maize (Zea mays L.), tomato (Solanum lycopersicum), grapes (Vitis vinifera), and other plants. The numbers of ARF genes that have been reported in various species are summarized in Table 1. ARFs were identified as large gene families and showed high homology in the same species. As a key factor in plant auxin signaling, ARFs can specifically bind to the auxin response element (AuxRE) in the promoter region of the auxin response genes and activate or repress the expression of auxin response gene, which in turn triggers many auxin-mediated physiological effects.
Table 1. Summary of the number of ARF genes in species that have been reported.
| Species | Gene No. |
|---|---|
| Agave americana | 32 |
| Ananas comosus | 20 |
| Arabidopsis thaliana | 23 |
| Aquilegia caerulea | 12 |
| Banana | 47 |
| Beta vulgaris | 16 |
| Boehmeria nivea | 14 |
| Brachypodium distachyon | 24 |
| Brassica rapa | 31 |
| Capsicum annuum | 22 |
| Carica papaya | 15 |
| Cicer arietinum | 28 |
| Citrus clementina | 17 |
| Citrus sinensis | 22 |
| Cucumis melo | 17 |
| Cucumis sativus | 15 |
| Dendrobium officinale | 14 |
| Dimocarpus longan | 17 |
| Eucalyptus grandis | 17 |
| Fagopyrum tataricum | 20 |
| Fragaria vesca | 17 |
| Glycine max | 55 |
| Gossypium raimondii | 35 |
| Hordeum vulgare | 20 |
| Jatropha curcas | 17 |
| Litchi chinensis | 39 |
| Malus domestica | 33 |
| Manihot esculenta | 18 |
| Marchantia polymorpha | 3 |
| Medicago truncatula | 24 |
| Mimulus guttatus | 19 |
| Mulberry | 17 |
| Oryza sativa | 25 |
| Osmanthus fragrans | 50 |
| Phaseolus vulgaris | 25 |
| Phyllostachys edulis | 24 |
| Physcomitrella patens | 12 |
| Populus trichocarpa | 39 |
| Prunus mume | 17 |
| Prunus persica | 17 |
| Prunus sibirica | 14 |
| Rafflesia cantleyi | 9 |
| Ricinus communis | 17 |
| Selaginella moellendorffii | 7 |
| Setaria italica | 23 |
| Solanum lycopersicum | 22 |
| Solanum tuberosum | 20 |
| Sorghum bicolor | 25 |
| Tamarix chinensis | 12 |
| Theobroma cacao | 19 |
| Triticum aestivum | 27 |
| Vitis vinifera | 19 |
| Zea mays | 36 |
| Ziziphus jujuba | 16 |
ARFs are structurally similar, with most members containing three regions: DBD (DNA-binding domain), MR (middle region), and PB1 (Phox and Bem 1). As shown in Figure 1, DBD is a DNA-binding domain directly involved in binding to AuxRE elements, while the MR region is responsible for activation or repression of target genes. It is believed that the amino acid composition and sequence length of MR region determine whether activation or inhibition. If this region is rich in proline, serine, and threonine it is able to produce inhibition, while activator-class ARFs often have glutamine-rich and leucine-rich MR regions. The PB1 domain can bind to Aux/IAA proteins to form heterodimers [1][2]. Some reports show that Aux/IAA and ARFs can form not only dimers but also larger complexes (oligomers), noting that oligomerization of Aux/IAA proteins may be essential for the inhibition of ARF proteins and only sufficient amounts of Aux/IAA proteins can exert the inhibitory effect of ARF proteins. The PB1 domain contributes to the oligomerization of ARFs and is important for Aux/IAA-ARF-mediated gene regulation [3][4]. The mode of action of ARF in auxin signaling is shown in Figure 1: When the cellular auxin concentration is low, Aux/IAA, encoded by the auxin early response gene family, binds to ARF and inhibits the activity of ARFs. When the auxin concentration is increased to a certain level, ubiquitinated Aux/IAA proteins are rapidly hydrolyzed. Aux/IAA protein’s inhibition of ARF transcription factors is relieved, and ARFs then promote or repress the expression of downstream genes.
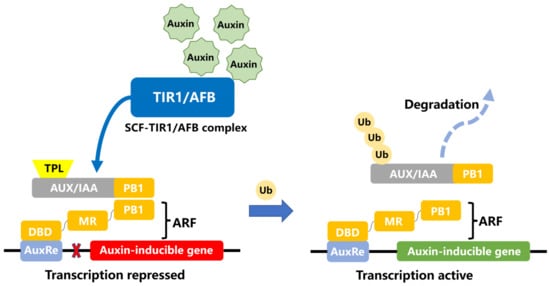
Figure 1. Model of ARF involvement in auxin response. The process of auxin signaling is coordinated by the auxin receptor TIR1/AFB, as well as the signal response factors AUX/IAA and ARF, which together mediate the auxin signaling response. Auxin acts as a “molecular glue” and facilitates the interaction between SCF-TIR1/AFB and Aux/IAA proteins. At low concentrations of auxin, the AUX/IAA repressor binds to the ARF transcription factor and forms a dimer that recruits the co-repressor TPL (TOPLESS) to inhibit the ARF activity and the expression of auxin responsive genes. When the concentration of auxin is increased, Aux/IAA binds to the SCF TIR1/AFB complex and is ubiquitinated and then degraded by 26S protease, and then ARF transcription factors are released to activate the transcription of downstream genes. DBD, DNA-binding domain; MR, middle region; PB1, Phox and Bem 1.
ARF is commonly expressed in all periods of plant growth and development and in different parts of the plant, which also determines its comprehensive influence on plant life activities. In addition, Estrada-Johnson et al. [5] found the gene family of strawberry (Fragaria ananassa) auxin response factors (FaARFs) were differentially expressed in the receptacle at four developmental stages by RNAseq. A large number of studies have shown that ARF is involved in the regulation of a variety of plant life activities, including leaf, flower, root development, and fruit ripening, senescence, and shedding. ARF is also involved in stress response and anthocyanin metabolism in plants. In addition, ARFs are also involved in plant hormone crosstalk, and the relevant research results are valuable for us to fully understand the regulatory effects of plant hormones. In recent years, studies on the regulation of plant life through hormone signaling pathways have been carried out, and increasingly more genome-wide identification and expression analyses of ARF gene families in plants have been reported, as shown in Table 1, which are of great significance for a more comprehensive understanding of the functions of ARF.
2. ARF Is Involved in Regulating Leaf Development
Most plants can perform photosynthesis by themselves, absorb carbon dioxide and release oxygen at the same time, and synthesize organic substances. This process is the prerequisite for all organisms on the earth to obtain energy. Leaves are the most important place for photosynthesis of plants. Important indicators such as the shape, size, and chlorophyll content of plant leaves usually determine whether photosynthesis can proceed normally and play a key role in the efficiency of photosynthesis [6]. In addition, in the leaves of plants, there are usually many other physiological activities, such as respiration. Generally speaking, there are stomata on the back of the leaves to support high-intensity respiration. Plants mainly maintain their own metabolic balance through these two activities on the leaves to maintain vitality. In addition, the transpiration also requires the participation of leaves.
At present, there have been a large number of reports showing the plant auxin plays a critical role in the life activity of leaves. A moderate increase in the concentration of auxin will stimulate the growth of leaves, while exceeding the optimal concentration will have an inhibitory effect. Auxin response factor (ARF), as a key regulator in the process of auxin signal transduction, directly participates in the regulation process of auxin on plant leaves in many links [7][8]. Arabidopsis thaliana is one of the first species to be used to study the regulation of ARF. In Arabidopsis, AtARF is already a well-studied member of the ARF family, and its regulatory role in leaves has been widely reported. AtARF2, AtARF3, AtARF4, and MONOPTEROS (MP)/ARF5 are involved in the regulation of auxin-responsive gene expression and leaf development. MP can promote the expression of WUSCHEL-RELATED HOMEOBOX1 (WOX1) and PRESSED FLOWER (PRS) directly, promoting leaf flattening. However, the expression of WOX1 and PRS can be repressed by several other members of the ARF family, AtARF2/3/4, as shown in Figure 2. These ARFs are all able to bind directly to the promoters of WOX1 and PRS [6]. Schuetz et al. [7] reported that ARF7 and ARF3 act synergistically with ARF5 in leaf formation, indicating a synergistic interaction, and these gene combinations work in parallel to maintain meristem organization and induce leaf formation. In addition, some studies have indicated that auxin induces leaf expansion by activating NPH4/ARF7 and ARF19 [8] (Figure 2). In tomato, Wu et al. [9] found that tomato leaf development was co-regulated by several members of the ARF family, including SlARF6A/8A/8B/24. In addition, SlARF6A/24 can influence the process of leaflet initiation by binding to the promoter of PIN-FORMED1 (SlPIN1), which affects the local accumulation of auxin. There are also reports that the chlorophyll content in leaves and fruit is significantly increased in tomato mutant lines of overexpressing SlARF6A. While downregulation of SlARF6A reduces the chlorophyll content [10]. ARF also plays a significant role in the leaves of some other species. In rice, OsARF19 controls rice leaf angles by binding to the promoters of the auxin early response gene OsGH3-5 and brassinosteroid (BR) response gene OsBRI1 [11].
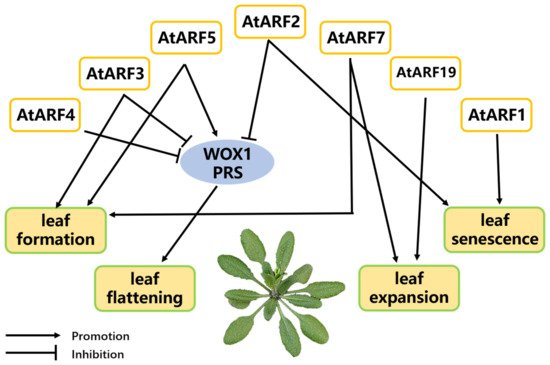
Figure 2. A model of ARFs involved in leaf development and senescence in Arabidopsis thaliana. ARF2/3/4/5 regulates leaf flattening by targeting the promoters of WUSCHEL-RELATED HOMEOBOX1 (WOX1) and PRESSED FLOWER (PRS). ARF3/5/7 synergistically regulate the leaf formation process. ARF 7 and 19 affect leaf expansion. ARF1 and 2 promote leaf senescence.
Leaf senescence is a necessary process of plant metabolism, accompanied by physiological changes such as the decrease of ribonucleic acid content, the destruction of chloroplasts, and the decrease of photosynthesis capacity. ARF also plays a regulatory role in the process. The AtARF1 and AtARF2 genes in Arabidopsis are involved in regulating leaf senescence, which is independent of the ethylene and cytokinin pathways [12][13]. In tomato, overexpression of SlARF2 accelerates leaf senescence [14]. In litchi, Zhang et al. [15] concluded that LcARF2 may be related to the senescence of litchi leaves.
3. ARF Is Involved in Regulating Root Development
Roots are the nutritional organs of plants, usually located below the ground surface, and are responsible for absorbing water and dissolving inorganic salts from the soil, as well as supporting, reproducing, and storing and synthesizing organic matter. Wang et al. [16] reported that microRNA160-targeted AtARF10/AtARF16 plays a crucial role in root cap formation. In rice, OsARF1 is also able to regulate crown roots [17]. Another ARF of rice species is OsARF12, which plays an active role in promoting root elongation [18].
The formation of lateral roots plays a key role in plant development, as well as the ability of plants to obtain nutrients depends on the degree of root branching. Numerous studies have suggested that auxin is a common signal for the formation of lateral roots. In Arabidopsis, AtARF5 is essential for lateral root formation [19]. In the root tissue of seedlings, AtARF7 and AtARF19 are transcriptional activators of early auxin response genes and control the expression of most plant auxin response genes. They regulate lateral root formation through direct activation of LBD/ASLs, as well as AtARF7 and AtARF19 double mutations caused impaired hypocotyl and root development [20]. In tomato, SlARF2 regulates the formation of lateral roots, and high expression of SlARF2 promotes lateral root growth [14].
In addition to lateral roots, ARF genes also play an important role in adventitious root development. AtARF6 and AtARF8 genes regulate adventitious root formation with the involvement of micro160 and micro167, and AtARF6 has been shown to positively control the development of adventitious roots. It has also been shown that AtARF17 is involved in this process and negatively regulates adventitious roots in Arabidopsis [21]. In rice, crown rootless1 (OsCRL1) encodes a plant-specific protein, and OsARF1 binds directly to AuxRE in the OsCRL1 promoter, which plays an important role in adventitious root initiation [22]. In addition, Yang et al. [23] reported PeARFs may play diverse regulatory roles in adventitious root development of Populus.
4. ARF Is Involved in the Regulation of Floral Structure and Sexual Reproduction
A plant’s flowers are essential for its own reproduction and are the organs of sexual reproduction for the vegetation. Over the course of evolution, the flower of a plant has developed into a more stable structure, generally consisting of a stalk, calyx, receptacle, corolla, pistil, and stamens. The stamens and pistils are composed of different parts, with the stamens consisting of the anthers and filaments and the pistils consisting of the style, stigma, and ovary. Flower development, maintenance, abscission process, and the normal formation of pistil and stamen are all crucial for plant growth and fruit formation. It has been extensively reported that auxin response factors play a key regulatory role in these processes.
Research on the regulatory role of ARF in plant flowers has been carried out mostly in Arabidopsis. Ellis et al. [12] reported that AtARF1 and AtARF2 in Arabidopsis affect flower initiation, stamen development, and flower senescence and abscission. arf2 mutant plants show delays in flowering and abscission of floral organs. arf1 mutations enhanced many arf2 phenotypes, indicating that ARF1 acts in a partially redundant manner with ARF2. AtARF3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling, and loss of AtARF3 leads to the development of pistil appearance defects [24]. There are many reports about the crucial roles of MONOPTEROS (MP)/ARF5 in the regulation of flower formation and other activities of shoot apical meristem. For example, Yamaguchi et al. [25] uncovered a molecular framework for flower initiation that includes AtARF5 and LEAFY, a master regulator of flower development, which reveals a mechanistic link between flower primordium initiation and subsequent steps in flower morphogenesis. AtARF6 and AtARF8 are thought to control stamen and pistil maturation, respectively, and the loss of a single gene in either ARF6 or ARF8 causes delayed stamen filament elongation and delayed anther dehiscence, resulting in reduced self-pollination.
5. ARF Is Involved in Regulating Fruit Development and Ripening
Research on fruit development and ripening has always been a hot topic. Human demand for high-quality fruits drives researchers to conduct in-depth research on the formation, development, and quality improvement of fruits, and its economic value is also huge. Plant fruits generally include two parts: pericarp and seed. Pericarp can be divided into exocarp, mesocarp, and endocarp, and seeds play a key role in the process of propagation and reproduction. The process of fruit development depends on cell division and cell expansion. After the completion of the pollination and ovary fertilization process, the carpel tissue around the ovary begins to expand rapidly [26]. The research on fruit includes the entire process from the occurrence of flowers to the later fruit preservation. For a long time, it has been believed that plant hormones play an important regulatory role in fruit growth and development, especially auxin, ethylene, and abscisic acid.
Some progress has been made in recent years regarding the role of ARF in the regulation of tomato fruit development and ripening. Figure 3 shows a model of ARFs regulating tomato fruit development and ripening. SlARF2 is an important part of tomato maturation regulatory network. Overexpression of SlARF2 enhances the expression of ethylene biosynthesis genes, increasing ethylene production and accelerating fruit ripening. However, downregulation or silencing of SlARF2 will lead to maturation defects and even inhibit maturity [27][28]. SlARF4 affects many physiological indicators of tomato fruits. For example, GOLDEN2-LIKE (GLK) transcription factors are required for the regulation of chloroplast and chlorophyll levels [29], and SlARF4 may control the accumulation of chlorophyll and the structure of the fruit cell wall through transcriptional inhibition of GLK1 expression in tomato fruits, as shown in Figure 3. Chlorophyll content is a key feature of immature fruit that affects the nutrient composition and flavor of mature fruit. Inhibition of SlARF4 expression can make tomato fruits show a dark green immature phenotype, and the resulting fruit is harder than the wild type. In addition, SlARF4 is also a major participant in mediating the control of auxin on sugar metabolism [30]. SIARF10 plays a similar role to SlARF4 and regulates chlorophyll and sugar accumulation during tomato fruit development through transcriptional activation of SlGLK1 [31]. SlARF10 is also involved in seed development and the regulation of fruit size and shape [32]. Another auxin response factor that has an effect on the chlorophyll content of tomato fruits is SlARF6A. The chlorophyll content in fruit was significantly increased in tomato mutant with overexpression of SlARF6A. In addition, ethylene production was inhibited, and fruit ripening rate decreased [10]. When pollination occurs, SlARF5 plays a role in fruit germination [33]. SlARF7 negatively regulates both fruit set and fruit development, slowing the plant auxin response during fruit growth [34][35]. In addition, SlARF8 and SlARF9 in tomato play a role in regulating fruit development and growth, and in negatively regulating cell division during early fruit development, respectively [31][36].
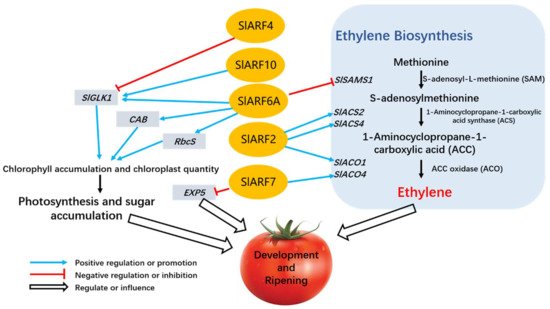
Figure 3. A model of ARFs regulating tomato fruit development and ripening. Several members of the auxin responsive factor family influenced the development and ripening of tomato fruits by regulating the expression of genes related to ethylene biosynthesis and chlorophyll accumulation. SlARF2 regulates ethylene synthesis and tomato ripening by regulating the expression of ethylene biosynthesis genes ACO1/ACS2/ACS4. GOLDEN2-LIKE transcription factor SlGLK1 plays an important role in chloroplast formation and chlorophyll accumulation. SlARF4 inhibits the expression of its encoding gene SlGLK1, while SlARF10 and SlARF6A promotes its expression. Moreover, SlARF6A also targets the promoters of CAB (genes encoding chlorophyll A/B binding) and RbcS (gene encoding ribulose bisphosphate carboxylase small chain), genes that affect chlorophyll accumulation, and positively regulates the expression of these genes. SlARF6A is also a negative regulator of SAMS1, which in turn negatively regulates ripening and ethylene biosynthesis in tomato fruit. EXPANSIN5 (EXP5) likely promotes cell expansion during fruit development. SlARF7 inhibits tomato fruit set and early development by directly regulating EXP5 and ACO4.
References
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574.
- Salehin, M.; Bagchi, R.; Estelle, M. SCFTIR1/AFB-Based Auxin Perception: Mechanism and Role in Plant Growth and Development. Plant Cell 2015, 27, 9–19.
- Korasick, D.A.; Westfall, C.S.; Lee, S.G.; Nanao, M.H.; Dumas, R.; Hagen, G.; Guilfoyle, T.J.; Jez, J.M.; Strader, L.C. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. USA 2014, 111, 5427–5432.
- Kato, H.; Mutte, S.K.; Suzuki, H.; Crespo, I.; Das, S.; Radoeva, T.; Fontana, M.; Yoshitake, Y.; Hainiwa, E.; van den Berg, W.; et al. Design principles of a minimal auxin response system. Nat Plants 2020, 6, 473.
- Estrada-Johnson, E.; Csukasi, F.; Pizarro, C.M.; Vallarino, J.G.; Kiryakova, Y.; Vioque, A.; Brumos, J.; Medina-Escobar, N.; Botella, M.A.; Alonso, J.M.; et al. Transcriptomic Analysis in Strawberry Fruits Reveals Active Auxin Biosynthesis and Signaling in the Ripe Receptacle. Front. Plant Sci. 2017, 8, 889.
- Guan, C.M.; Wu, B.B.; Yu, T.; Wang, Q.Q.; Krogan, N.T.; Liu, X.G.; Jiao, Y.L. Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis. Curr. Biol. 2017, 27, 2940–2950.
- Schuetz, M.; Fidanza, M.; Mattsson, J. Identification of Auxin Response Factor-Encoding Genes Expressed in Distinct Phases of Leaf Vein Development and with Overlapping Functions in Leaf Formation. Plants 2019, 8, 242.
- Wilmoth, J.C.; Wang, S.C.; Tiwari, S.B.; Joshi, A.D.; Hagen, G.; Guilfoyle, T.J.; Alonso, J.M.; Ecker, J.R.; Reed, J.W. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005, 43, 118–130.
- Wu, L.J.; Tian, Z.D.; Zhang, J.H. Functional Dissection of Auxin Response Factors in Regulating Tomato Leaf Shape Development. Front. Plant Sci. 2018, 9, 957.
- Yuan, Y.J.; Xu, X.; Gong, Z.H.; Tang, Y.W.; Wu, M.B.; Yan, F.; Zhang, X.L.; Zhang, Q.; Yang, F.Q.; Hu, X.W.; et al. Auxin response factor 6A regulates photosynthesis, sugar accumulation, and fruit development in tomato. Hortic. Res. 2019, 6, 1–16.
- Zhang, H.H.; Li, L.L.; He, Y.Q.; Qin, Q.Q.; Chen, C.H.; Wei, Z.Y.; Tan, X.X.; Xie, K.L.; Zhang, R.F.; Hong, G.J.; et al. Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA viruses for infection. Proc. Natl. Acad. Sci. USA 2020, 117, 9112–9121.
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574.
- Wang, S.X.; Shi, F.Y.; Dong, X.X.; Li, Y.X.; Zhang, Z.H.; Li, H. Genome-wide identification and expression analysis of auxin response factor (ARF) gene family in strawberry (Fragaria vesca). J. Integr. Agr. 2019, 18, 1587–1603.
- Rena, Z.; Liu, R.; Gu, W.; Dong, X. The Solanum lycopersicum auxin response factor SlARF2 participates in regulating lateral root formation and flower organ senescence. Plant Sci. 2017, 256, 103–111.
- Zhang, Y.Q.; Zeng, Z.H.; Chen, C.J.; Li, C.Q.; Xia, R.; Li, J.G. Genome-wide characterization of the auxin response factor (ARF) gene family of litchi (Litchi chinensis Sonn.): Phylogenetic analysis, miRNA regulation and expression changes during fruit abscission. Peerj 2019, 7, e6677.
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216.
- Waller, F.; Furuya, M.; Nick, P. OsARF1, an auxin response factor from rice, is auxin-regulated and classifies as a primary auxin responsive gene. Plant Mol. Biol. 2002, 50, 415–425.
- Qi, Y.H.; Wang, S.K.; Shen, C.J.; Zhang, S.N.; Chen, Y.; Xu, Y.X.; Liu, Y.; Wu, Y.R.; Jiang, D.A. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 2012, 193, 109–120.
- Goh, T.; Kasahara, H.; Mimura, T.; Kamiya, Y.; Fukaki, H. Multiple AUX/IAA-ARF modules regulate lateral root formation: The role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1461–1468.
- Kalve, S.; Sizani, B.L.; Markakis, M.N.; Helsmoortel, C.; Vandeweyer, G.; Laukens, K.; Sommen, M.; Naulaerts, S.; Vissenberg, K.; Prinsen, E.; et al. Osmotic stress inhibits leaf growth of Arabidopsis thaliana by enhancing ARF-mediated auxin responses. New Phytol. 2020, 226, 1766–1780.
- Mallory, A.C.; Bartel, D.P.; Bartel, B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 2005, 17, 1360–1375.
- Inukai, Y.; Sakamoto, T.; Ueguchi-Tanaka, M.; Shibata, Y.; Gomi, K.; Umemura, I.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 2005, 17, 1387–1396.
- Yang, C.X.; Xu, M.; Xuan, L.; Jiang, X.M.; Huang, M.R. Identification and expression analysis of twenty ARF genes in Populus. Gene 2014, 544, 134–144.
- Zhang, K.; Wang, R.Z.; Zi, H.L.; Li, Y.P.; Cao, X.W.; Li, D.M.; Guo, L.; Tong, J.H.; Pan, Y.Y.; Jiao, Y.L.; et al. AUXIN RESPONSE FACTOR3 Regulates Floral Meristem Determinacy by Repressing Cytokinin Biosynthesis and Signaling. Plant Cell 2018, 30, 324–346.
- Yamaguchi, N.; Wu, M.F.; Winter, C.M.; Berns, M.C.; Nole-Wilson, S.; Yamaguchi, A.; Coupland, G.; Krizek, B.A.; Wagner, D. A Molecular Framework for Auxin-Mediated Initiation of Flower Primordia. Dev. Cell 2013, 24, 271–282.
- Pattison, R.J.; Csukasi, F.; Catala, C. Mechanisms regulating auxin action during fruit development. Physiol. Plant. 2014, 151, 62–72.
- Breitel, D.A.; Chappell-Maor, L.; Meir, S.; Panizel, I.; Puig, C.P.; Hao, Y.W.; Yifhar, T.; Yasuor, H.; Zouine, M.; Bouzayen, M.; et al. AUXIN RESPONSE FACTOR 2 Intersects Hormonal Signals in the Regulation of Tomato Fruit Ripening. PLoS Genet. 2016, 12, e1005903.
- Hao, Y.W.; Hu, G.J.; Breitel, D.; Liu, M.C.; Mila, I.; Frasse, P.; Fu, Y.Y.; Aharoni, A.; Bouzayen, M.; Zouine, M. Auxin Response Factor SlARF2 Is an Essential Component of the Regulatory Mechanism Controlling Fruit Ripening in Tomato. PLoS Genet. 2015, 11, e1005649.
- Waters, M.T.; Moylan, E.C.; Langdale, J.A. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 2008, 56, 432–444.
- Sagar, M.; Chervin, C.; Roustan, J.P. Under-expression of the Auxin Response Factor Sl-ARF4 improves postharvest behavior of tomato fruits. Plant Signal. Behav. 2013, 8, e25647.
- Yuan, Y.J.; Mei, L.H.; Wu, M.B.; Wei, W.; Shan, W.; Gong, Z.H.; Zhang, Q.; Yang, F.Q.; Yan, F.; Zhang, Q.; et al. SIARF10, an auxin response factor, is involved in chlorophyll and sugar accumulation during tomato fruit development. J. Exp. Bot. 2018, 69, 5507–5518.
- Hendelman, A.; Buxdorf, K.; Stav, R.; Kravchik, M.; Arazi, T. Inhibition of lamina outgrowth following Solanum lycopersicum AUXIN RESPONSE FACTOR 10 (SlARF10) derepression. Plant Mol. Biol. 2012, 78, 561–576.
- Liu, S.Y.; Zhang, Y.W.; Feng, Q.S.; Qin, L.; Pan, C.T.; Lamin-Samu, A.T.; Lu, G. Tomato AUXIN RESPONSE FACTOR 5 regulates fruit set and development via the mediation of auxin and gibberellin signaling. Sci. Rep. 2018, 8, 2971.
- De Jong, M.; Wolters-Arts, M.; Feron, R.; Mariani, C.; Vriezen, W.H. The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 2009, 57, 160–170.
- Hu, J.H.; Israeli, A.; Ori, N.; Sun, T.P. The Interaction between DELLA and ARF/IAA Mediates Crosstalk between Gibberellin and Auxin Signaling to Control Fruit Initiation in Tomato. Plant Cell 2018, 30, 1710–1728.
- De Jong, M.; Wolters-Arts, M.; Schimmel, B.C.J.; Stultiens, C.L.M.; de Groot, P.F.M.; Powers, S.J.; Tikunov, Y.M.; Bovy, A.G.; Mariani, C.; Vriezen, W.H.; et al. Solanum lycopersicum AUXIN RESPONSE FACTOR 9 regulates cell division activity during early tomato fruit development. J. Exp. Bot. 2015, 66, 3405–3416.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.6K
Revisions:
2 times
(View History)
Update Date:
15 Feb 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




