You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Abdul Razak Mariatulqabtiah | + 3727 word(s) | 3727 | 2022-01-06 05:08:58 | | | |
| 2 | Dean Liu | Meta information modification | 3727 | 2022-02-14 07:40:05 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Mariatulqabtiah, A.R. Loop-Mediated Isothermal Amplification. Encyclopedia. Available online: https://encyclopedia.pub/entry/19317 (accessed on 22 December 2025).
Mariatulqabtiah AR. Loop-Mediated Isothermal Amplification. Encyclopedia. Available at: https://encyclopedia.pub/entry/19317. Accessed December 22, 2025.
Mariatulqabtiah, Abdul Razak. "Loop-Mediated Isothermal Amplification" Encyclopedia, https://encyclopedia.pub/entry/19317 (accessed December 22, 2025).
Mariatulqabtiah, A.R. (2022, February 10). Loop-Mediated Isothermal Amplification. In Encyclopedia. https://encyclopedia.pub/entry/19317
Mariatulqabtiah, Abdul Razak. "Loop-Mediated Isothermal Amplification." Encyclopedia. Web. 10 February, 2022.
Copy Citation
The loop-mediated isothermal amplification method (LAMP) is distinguished by the utilization of at least four different primers which specifically recognize six distinct regions on the target nucleotide sequence.
avian
virus
diagnostic
LAMP
1. Introduction
The loop-mediated isothermal amplification method (LAMP) is distinguished by the utilization of at least four different primers which specifically recognize six distinct regions on the target nucleotide sequence [1]. The LAMP reaction is facilitated by unique Bst polymerase with strand displacement activity which does not require denaturation of the double-stranded DNA during the amplification process. Since the heating of nucleic during the denaturation step is inessential, reaction can proceed optimally at a constant temperature of around 60 °C to 70 °C in a single step reaction [2]. While the alignment of the LAMP primers at six target regions in one reaction further increases the specificity of the assay, the reaction of this method is presented to be fast and much simpler without reliance on expensive equipment. Using the LAMP method, positive results can be recorded in as little as 20 to 30 min in a single-step reaction [1][3]. The detection of RNA can also be achieved by adding reverse transcriptase to the reaction, or using isothermal polymerase with reverse transcription activity for reverse-transcription LAMP (RT-LAMP).
2. Origin and Advancements of the LAMP Method
The discovery of the LAMP method by Notomi et al. [1] dates back more than two decades, and thereafter was patented by Eiken Chemical Co., Ltd. in Tokyo, Japan [3]. The gene amplification method was developed primarily based on the application of the uniquely designed primers that function together in a set. The main LAMP primers consist of a forward outer primer and backward outer primer called F3 and B3, respectively, and the key binding primers that form the stem loops, creating a structure called the forward inner primer (FIP) and backward inner primer (BIP), as depicted in Figure 1. The optional addition of loop primer forward (LF) and loop primer backward (LB) serve as starting materials to enhance the DNA synthesis, and thus reduce the amplification time [4]. The duration of the LAMP reaction can be further shortened by the inclusion of a forward swarm primer and backward swarm primer that target the upstream region of the FIP and BIP binding sequences [5].
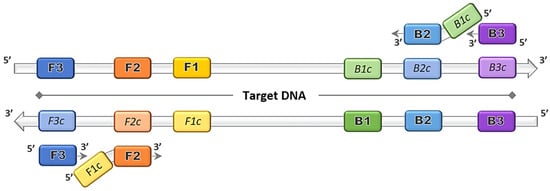
Figure 1. Loop-mediated isothermal amplification (LAMP) primers binding on target DNA. Four principal LAMP primers namely F3, FIP (F2 + F1c), B3 and BIP (B2 + B1c) bind on the complementary target regions and amplifications proceed in the direction of 5′ to 3′.
Prior to the LAMP primer design, any target sequence should be analyzed to determine the region to which the primer set would align; either to a specific strain or a conserved sequence within closely related species. Multiple sequence alignment (MSA) of available genomes or published sequences identifies conserved regions, which serve as the template for the primer design. A concise LAMP primer design is crucial to generate a set of functional primers that would perform at optimum level under the feasible conditions. The features that should be taken into measure are oligonucleotide length, compositions of guanine and cytosine (G-C content), melting temperature and the stability of the primer ends. For the LAMP primers, F3 and B3 should have a length of 18 to 20 nucleotides, while both FIP and BIP are set to have a functional length between 38 and 42 nucleotides [1]. The G-C content LAMP primer set would recognize a target region of within 300 nucleotides, short enough to detect small conserved regions in many genes. The GC content in LAMP oligonucleotides, like most primers, is selected to be between 50% and 60% for GC-normal and GC-rich regions. For the melting temperature (Tm) of the oligonucleotides, the value can be predicted using the nearest neighbour method to obtain a value around 55 °C to 60 °C for an AT-rich region and 60 °C to 65 °C GC-rich region. Another key feature in LAMP primer design is the stability of the oligonucleotide ends. The LAMP primer stability is calculated via the delta G value, in coherence with the standard PCR primer design to be less than −4 kcal/mol [6].
While each parameter in developing the primers needs to be addressed precisely, the design steps have been simplified using the available online software. The first and most widely used online software dedicated to LAMP primer design is Primer Explorer (https://primerexplorer.jp/e/ accessed on 1 September 2021), developed by Eiken Chemical Co., Ltd. (Tokyo, Japan). The free-access software can generate multiple sets of LAMP primers based on the input target sequence with automated yet adjustable primer designing parameters. Besides, several LAMP-designing primer software packages have also been created to ease research in developing the LAMP method for a wide range of applications;
3. LAMP Application in Screening of Avian Viruses
3.1. Detection of Avian Influenza Viruses of Multiple Subtypes
Avian influenza viruses (Influenza A virus) are the highly infectious viruses that have affected the poultry industries for many years. The highly pathogenic avian influenza (HPAI) virus has caused severe economic damage to the poultry industry with up to 100% mortality. For example, the outbreak of avian influenza H7N1 in Italy caused more than 13 million chicken deaths from 1999 to 2000 [7]. The affected hosts are not only food producing birds such as the chicken, quail and turkey, but also pet birds and wild birds. As a result of high vulnerability and mortality in chickens due to the HPAIs, monitoring and screening against such viruses are crucial to secure the essential poultry source and control the spread of the avian influenza diseases towards susceptible environments. Sporadic cases of zoonotic infection of animal-to-human transmission that ended up with mortality have also been reportedly caused by avian influenza virus subtypes H1N1, H2N2, H5N1, H7N7 and H7N9 [8][9].
Influenza A virus is the only species that belongs to the genus of Alphainfluenzavirus, with major natural reservoirs in birds and some mammals [10]. Many subtypes of the Influenza A virus have been discovered, and classifications are made according to combinations of the different viral glycoproteins hemagglutinin (H) and neuraminidase (N). Among the highly pathogenic subtypes are H5 and H7 [7]. These RNA viruses have a total of around 13.5 kb of genome that are divided into eight RNA segments. The efforts in screening of influenza A viruses have been crucial to control the spread of the pathogens while mitigating the losses caused. Together with conventional and sensitive screening methods such as qPCR, RT-qPCR, nucleic acid sequence-based amplification (NASBA) and next-generation sequencing (NGS), the LAMP method has been applied for genetic detection of the highly pathogenic avian viruses [10].
The widespread avian influenza virus subtype H5N1 has been accurately detected using the LAMP method in many studies [8][7][11][12][13][14][15][16]. The detection of avian influenza virus subtype H7 using the LAMP method has shown a promising sensitivity at a level of 0.01 PFU/tube in the assay, which was 100-fold more sensitive than RT-PCR in a comparison study [7]. The sensitivity obtained in the H7-RT-LAMP method is in agreement with the research on LAMP diagnosis against avian influenza virus subtype H5 [12]. The detection limit of the LAMP method in detecting the LPAI H7 subtypes was also as good as that of HPAI [7]. Clinical samples of throat and cloacal swabs have higher predictive value in surveillance screening of highly pathogenic influenza viruses [7][11]. From a cultured specimen containing avian influenza virus subtype H9, the detection rate of confirmed positive samples using RT-LAMP was recorded at 100%, while RT-PCR had a lower positive rate at 91.8% due to several false negative results [11]. The detection limit of the H9 subtype was recorded at 10 copy numbers, which is tenfold more sensitive than that of RT-PCR [11]. The RT-LAMP method developed for the detection of avian influenza subtype H7N9 showed that the assay is highly specific with sensitivity of up to 50 copies/reaction and applicable for a direct RT-LAMP without the additional nucleic acid extraction step [8]. The high sensitivity level of the RT-LAMP method in the previous avian influenza studies indicates that this method is viable for the detection of the early stage or newly acquired infections with low viral amounts. Recently, a set of LAMP primers designed on the matrix (M) gene of Alphainfluenzavirus was able to screen all subtypes of avian influenza viruses, which included positive detection of subtypes H1, H5 and H9 from clinical specimens [15]. The broad-range LAMP method has recorded a faster detection time in all reactions as compared to the previous LAMP method targeting the matrix (M) gene [17].
3.2. Screening of Polyomaviruses in Birds
Polyomaviruses of birds under the genus Gammapolyomavirus or Avipolyomavirus consist of several species of virus that are host-specific to different species of birds. Among the recorded species of avian polyomaviruses in the virus taxonomy are budgerigar fledgling disease virus, Adelie penguin polyomavirus, butcherbird polyomavirus, canary polyomavirus, crow polyomavirus, finch polyomavirus and goose hemorrhagic polyomavirus [18][19][20][21]. Known to cause diseases and mortality in many species of birds, this group of avian polyomaviruses carry a circular genome of around 5 kb of double-stranded DNA in a non-encapsulated icosahedral capsid [19][22]. Pathological symptoms of birds infected with avian polyomaviruses, following clinical analysis and histopathologic lesions, include hepatitis, hydropericardium and ascites [18][23]. Fledgling or young birds are the main groups affected by avian polyomaviruses with high morbidity and mortality rates, and the positive cases are more prominent with birds kept in captivity with close proximity [23][24].
The incidences of goose hemorrhagic polyomavirus (GHPyV) infection have caused economic losses in waterfowl production at large scale, with a 32% mortality rate of young goslings reported in Poland [21][25]. Adult parrots infected with budgerigar fledgling disease virus (Aves polyomavirus 1, APyV) are more resistant to the inflammatory diseases and while they appear to be asymptomatic, the virus can be shed via feathers and faeces for up to 6 months [26][27]. The fact that the virus can survive in an outside environment for many months raises concerns for swift yet efficient measures such as early screening, cage sanitization and isolation of the infected birds. Budgerigar fledgling disease virus is also observed to co-infect parrots together with beak and feather disease virus (BFDV) in many cases [28][29][30]. Hence a diagnostic method that is able to segregate the existence of different viruses is necessary to give specific treatments for the infected birds.
Among the developed diagnosis methods for the molecular detection of different avian polyomaviruses are LAMP, PCR, qPCR and enzyme-linked immunosorbent assay (ELISA) [21][24]. A study on the detection of goose hemorrhagic polyomavirus in geese and ducks showed the LAMP method has a good sensitivity in limit of detection test at 1.5 pg of extracted DNA, despite qPCR displaying 100-fold more sensitive results [21]. Positive detections were recorded from clinical specimens with diseased symptoms, indicating the consistency of the LAMP method in detecting the virus as confirmed by the qPCR results using the similar specimens [21]. To verify the possibility of any cross-reactivity, the optimized LAMP method was tested against GHPyV with other pathogens present in ducks and geese to observe the potential of cross-reactivity that might lead to a false positive result. Prevalent infections of budgerigar fledgling disease virus (APyV) among parrots are concerning, and should not be neglected in the pet bird industry as well as the global parrot population, which is why the LAMP method has been recently developed for efficient screening of this virus [26]. Using hydroxyl naphthol blue (HNB) in the reaction mixture for direct monitoring of the results, the colorimetric LAMP method on budgerigar fledgling disease virus has shown 100% sensitivity and specificity in diagnostic performance [26]. The exclusivity of the developed LAMP method was also assessed using different avian pathogens, including the BFDV that commonly co-infects parrots [26]. The limit of APyV detection was observed at 500 copies/reaction for both LAMP and PCR methods [26]. The analytical sensitivity results of LAMP being as great as, if not greater than, those from qPCR suggests that the LAMP method is satisfactory to replace the standard diagnostic as a handy and cheaper alternative, especially for diagnostic application in small veterinary clinics or on-field screening.
3.3. Diagnosis of Circoviruses in Different Avian Hosts
Circoviruses are among the cluster of small viruses in the Circoviridae family, with non-enveloped icosahedral virions of approximately 20 nm in diameter. Enclosed within the virions are circular genomes of single-stranded DNA, with sizes ranging from 1.8 kb to 2.2 kb according to the species that infect different mammalian hosts. Within the avian hosts, infections of circoviruses have been reported in the goose, canary, pigeon, swan, duck and parrot [31][32][33][34][35]. Circoviruses in avian hosts are identified to cause immunosuppression, making the hosts much more vulnerable to secondary infections and eventually leading to mortality [31][34][35].
Goose circovirus (GoCV) is often associated with runting and stunting syndromes in the commercial geese flock with a high degree of mortality [32][34]. Feather disorders are the non-specific clinical symptoms while pathological studies revealed hemorrhages and splenomegaly [34]. The virus also causes T-lymphocyte depletion in lymphoid organs, which results in an immunosuppressive effect [34]. Screening of this virus via the LAMP method offers early detection to swiftly isolate the infected birds from the remaining flock, especially when asymptomatic infections occur [32]. The simple diagnosis method was developed by specifically targeting the Vgp4 gene of the GoCV, which codes for one of the viral capsid proteins. The LAMP method on goose circovirus showed a detection limit at 100 pg DNA equal to the qPCR method, while no false positive was observed with DNA samples of other avian viruses infecting geese such as goose hemorrhagic polyomavirus or goose parvovirus [32]. Field samples with clinical symptoms were tested positive at 97.4% sensitivity using the optimized LAMP method in comparison study with qPCR assay [32].
As the name itself suggests, beak and feather disease virus (BFDV) has been identified to cause severe feather loss and abnormal beak shape as the significant symptoms of this circovirus infection in parrots, similar to the goose circovirus symptoms [31][36]. BFDV infection is the most common viral disease affecting parrots, highly transmittable horizontally and vertically to more than 60 different species of psittacine [37]. A LAMP study has demonstrated an excellent limit of detection against BFDV at 3.5 fg of viral DNA following optimal reaction temperature at 63 °C [31]. Clinical specimens of tissues and livers from parrots suspected to be infected with BFDV were tested with both conventional PCR and optimized LAMP methods, where the LAMP method showed greater sensitivity by detecting more positive samples [31]. The application of this simple sample pre-treatment method highlights the feasibility of LAMP in detecting target pathogens in different types of samples for a rapid diagnostic procedure. In a more recent development, the LAMP reaction has been incorporated with swarm primers to reduce the reaction time of BFDV detection to 40 min of complete reaction time and the results can be directly visualized with color changes of the hydroxy naphthol blue dye [38]. LAMP with swarm primers also demonstrated a great improvement in diagnostic performance by showing 100% clinical sensitivity from 71% of the standard LAMP method, following BFDV detection from psittacine samples of blood, feather, tissue and cloacal swabs [38]. Early and rapid diagnosis using the LAMP method to screen infected parrots prior to any international trade would help to control virus transmission [38].
3.4. Screening of Immunosuppressive Viruses in Chickens
The economically endangering chicken anemia virus (CAV), or Gyrovirus, was once grouped within the family of Circoviridae until a recent taxonomical classification was made to classify the virus into the family of Anelloviridae due to significant differences in genome organization [39][40]. As a sole species under the Gyrovirus genus, this single-stranded DNA virus has caused the infectious chicken anemia disease that affects the poultry industry worldwide [41]. Similar to the viruses from the Circoviridae family, CAV is known to cause immunosuppression of the host by replicating inside the cortex thymus to destroy the precursor T cells [40]. The immunosuppressed chickens are more likely to acquire secondary infections that increase morbidity and mortality, which ultimately cause economic loss. Another replication site of the CAV is in hemocytoblast of the bone marrow, where the destruction of these stem cells leads to aplastic anemia [40]. Epidemiological studies showed that CAV infects a majority of chicks as young as 1 day old, making them the most vulnerable group via the vertical transmission of hatching eggs [42]. Meanwhile horizontal transmissions can occur via the fecal–oral route and diseased feather follicle epithelium from the infected litter within flocks [40]. Complete eradication of CAV in contaminated cages is less likely due to the virus having high resistance to heat and chemical disinfectants [40]. While currently there is no proper medication to specifically treat CAV infection, a common approach in controlling the viral outbreak is through vaccination of the breeding chickens prior to the stage of egg production [40][42]. Immunological assays, conventional PCR and viral isolation are the diagnostic methods used to screen for the presence of CAV [42][43]. Though viral isolation is not often applied for diagnosis due to the meticulous procedures needed together with high cost, a novel LAMP detection of CAV was proposed as a simple and inexpensive alternative. The LAMP method targeting the VP2 gene of CAV was able to detect template DNA at a minimum amount of 100 fg within 30 min of reaction [42]. CAV DNA from the liver of an infected broiler chicken was successfully distinguished by the LAMP method, corresponding to the clinical result from the conventional PCR method [42]. This method can be adapted as a suitable tool for occasional surveillance and epidemiological studies of CAV infection.
Marek’s disease virus (MDV) or the scientific name Gallid herpesvirus 2, a pathogenic and oncogenic serotype 1 of the family Herpesviridae, is one of the most infectious pathogens in poultry production [44]. It was reported that ubiquitous MDV infections occurred frequently in more than 70 countries, including the emergence of very virulent strains (vvMDV+) [45]. Upon entry via inhalation of infected dusts, MDV is transmitted to B cells and T cells for viral replication to take place, which may cause cell death [46]. The infected cells reach latency phase to proliferate abnormally and spread within the host, causing tumors in the liver, spleen and kidney [44][46]. The vaccination programs for MDV have reported an increase in demand for the protection against this avian virus, including double dosage and revaccination, which highlights the urgency to tackle Marek’s disease globally [45]. Several studies have utilized the LAMP method in detecting the presence of MDV, where the findings highlight efficiency and accuracy of this simple method as the major advantages in handling viral diagnosis [43][44][47][48]. Clinical evaluation has shown a positive detection rate at 95%, with the limit of detection as low as 50 copies [43][47]. In a very practical method, the dust of fine skin particles and feather debris from chickens infected with MDV were able to produce positive detection using a simple heat treatment method [44].
Another immunosuppressive disease affecting the poultry industry is infectious bursal disease (IBD), instigating morbidity of almost 100% in susceptible flocks while very virulent strains are deadly enough at a mortality rate of around 60% [43][49]. IBD is also known as Gumboro disease and infectious bursitis, caused by the only species of the Avibirnavirus genus, namely infectious bursal disease virus (IBDV) [50]. This double-stranded RNA virus is most lethal towards young chicks of age between 3 to 6 weeks old at lower antibody levels [4][43]. Pathogenic Serotype 1 of IBDV destroys the B lymphocytes in the chicken’s bursa, impeding the host humoral immunity and consequently becoming more vulnerable to secondary infections by other pathogens [4]. RT-LAMP tested against the IBDV positive clinical samples demonstrated 100% sensitivity and specificity [4][43]. Using a biotin-labeled DNA probe designed to target the amplified RT-LAMP products, a lateral flow dipstick was applied for the molecular detection of IBDV [4]. FITC-labeled probes were added following the RT-LAMP reaction for the hybridization to occur with the DNA probes. The positive result could be visualized using naked eyes by either the turbidity of the white precipitate or the dark purple band developed on the test line of the dipstick [4].
3.5. Wide Application of LAMP against Notable Viruses in Poultry Industry
Apart from these highlighted avian viruses that have been successfully screened using the sensitive yet rapid LAMP detection, this method has been widely utilized in many other viruses that adversely affect the poultry industry. Such LAMP application includes distinguishing the subgroup of avian leukosis virus (ALV) from other exogenous Alpharetrovirus subgroups [51][52]. The ALV subgroups are segregated based on the glycoproteins of the viral envelope, a major factor that regulates antigenicity, host range and the occurrence of viral interference among the group [52]. While all the ALV field strains are oncogenic, the neoplastic lymphoid leukosis disease triggered by this cluster of RNA viruses on chicken flocks needs to be identified and addressed specifically according to the subgroup. Infectious bronchitis virus (IBV) is another major cause of respiratory disease in chicken farms that has been underlined with the urgency of rapid diagnosis via the optimized LAMP method [53][54][55]. With single-stranded RNA avian coronavirus as the causative agent, from the Coronaviridae family, IBV was first tested with RT-LAMP at the conserved regions of the nucleocapsid gene [53]. Through an innovative mRT-LAMP-LFD, a multiple LAMP method was coupled with a lateral flow dipstick for simultaneous detection of IBV and the contagious Newcastle disease virus (NDV) [55]. In the diagnostic tests, the presence of IBV and NDV on clinical samples detected using the mRT-LAMP-LFD assay were at decent sensitivity rates of 98.65% and 97.25%, respectively. Single-step RT-LAMP on IBV that relies on fluorescence emissions was developed to facilitate semi-quantification of the virus spread at molecular level in veterinary laboratories which lack mainstream instruments [54].
Besides chickens, ducks are also heavily affected by avian viruses in large-scale poultry production. Duck viral hepatitis (DVH) is a lethal and highly contagious disease typically affecting young ducklings. Known to be associated with substantial liver lesions with a concerning death rate, the most widespread viral strain is named duck hepatitis A virus type-1 (DHAV-1) from the Picornaviridae family. The presence of this RNA virus was distinguished through an established RT-LAMP method by targeting the conserved regions in the 3D gene, with a detection limit as low as 0.3 pg [56][57]. The RT-LAMP method was also used to detect Tembusu virus (TMUV), a species of Flavivirus virus that causes large economic loses in egg-laying and breeder duck farms, especially in China [58][59]. This virus species from the Flaviviridae family is associated with many forms of transmissions, such as mosquito borne, airborne, direct contact and vertical transmission, which could all be regulated through point-of-care detection at infected farms using the viable RT-LAMP method [58][60][61]. Advocated as a minimal technology analytical tool, routine screening in poultry farms would surely be of benefit to regulate any virus transmission to avian hosts.
Figure 3 summarizes the viruses (in short form) detected using the LAMP method as discussed throughout the content under different groups of the avian hosts; chicken, duck, goose and parrot.
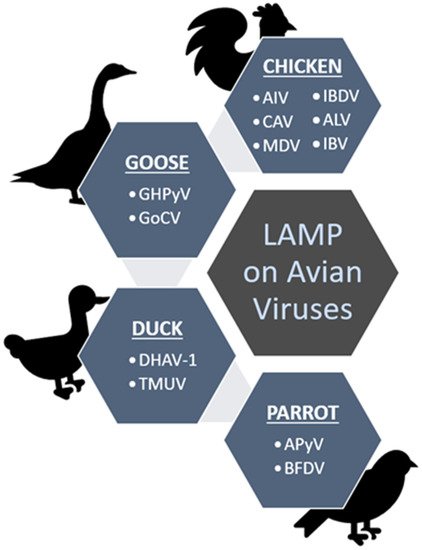
Figure 3. Summary of the discussed avian viruses that have been screened from chickens, geese, ducks and parrots using the established LAMP methods.
References
- Notomi, T.; Okayama, H.; Masubuchai, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63.
- Parida, M.; Sannarangaiah, S.; Dash, P.K.; Rao, P.V.L.; Morita, K. Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008, 18, 407–421.
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol. 2015, 53, 1–5.
- Tsai, S.-M.; Liu, H.-J.; Shien, J.-H.; Lee, L.-H.; Chang, P.-C.; Wang, C.-Y. Rapid and sensitive detection of infectious bursal disease virus by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick. J. Virol. Methods 2012, 181, 117–124.
- Martineau, R.L.; Murray, S.A.; Ci, S.; Gao, W.; Chao, S.-H.; Meldrum, D.R. Improved Performance of Loop-Mediated Isothermal Amplification Assays via Swarm Priming. Anal. Chem. 2017, 89, 625–632.
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134.
- Bao, H.; Wang, X.; Zhao, Y.; Sun, X.; Li, Y.; Xiong, Y.; Chen, H. Development of a reverse transcription loop-mediated isothermal amplification method for the rapid detection of avian influenza virus subtype H7. J. Virol. Methods 2012, 179, 33–37.
- Nakauchi, M.; Takayama, I.; Takahashi, H.; Tashiro, M.; Kageyama, T. Development of a reverse transcription loop-mediated isothermal amplification assay for the rapid diagnosis of avian influenza A (H7N9) virus infection. J. Virol. Methods 2014, 204, 101–104.
- Reperant, L.A.; Kuiken, T.; Osterhaus, A.D. Adaptive pathways of zoonotic influenza viruses: From exposure to establishment in humans. Vaccine 2012, 30, 4419–4434.
- Okamatsu, M.; Hiono, T.; Kida, H.; Sakoda, Y. Recent developments in the diagnosis of avian influenza. Vet. J. 2016, 215, 82–86.
- Chen, H.-T.; Zhang, J.; Sun, D.-H.; Ma, L.-N.; Liu, X.-T.; Cai, X.-P.; Liu, Y.-S. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of H9 avian influenza virus. J. Virol. Methods 2008, 151, 200–203.
- Imai, M.; Ninomiya, A.; Minekawa, H.; Notomi, T.; Ishizaki, T.; Tashiro, M.; Odagiri, T. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine 2006, 24, 6679–6682.
- Imai, M.; Ninomiya, A.; Minekawa, H.; Notomi, T.; Ishizaki, T.; Van Tu, P.; Tien, N.T.K.; Tashiro, M.; Odagiri, T. Rapid diagnosis of H5N1 avian influenza virus infection by newly developed influenza H5 hemagglutinin gene-specific loop-mediated isothermal amplification method. J. Virol. Methods 2007, 141, 173–180.
- Postel, A.; Letzel, T.; Frischmann, S.; Grund, C.; Beer, M.; Harder, T. Evaluation of Two Commercial Loop-Mediated Isothermal Amplification Assays for Detection of Avian Influenza H5 and H7 Hemagglutinin Genes. J. Vet. Diagn. Investig. 2010, 22, 61–66.
- Shi, L.; Yu, X.-W.; Yao, W.; Yu, B.-L.; He, L.-K.; Gao, Y.; Zhang, Y.-X.; Tian, G.-B.; Ping, J.-H.; Wang, X.-R. Development of a reverse-transcription loop-mediated isothermal amplification assay to detect avian influenza viruses in clinical specimens. J. Integr. Agric. 2019, 18, 1428–1435.
- Yoshida, H.; Sakoda, Y.; Endo, M.; Motoshima, M.; Yoshino, F.; Yamamoto, N.; Okamatsu, M.; Soejima, T.; Senba, S.; Kanda, H.; et al. Evaluation of the Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) as a Screening Method for the Detection of Influenza Viruses in the Fecal Materials of Water Birds. J. Vet. Med. Sci. 2011, 73, 753–758.
- Shivakoti, S.; Ito, H.; Murase, T.; Ono, E.; Takakuwa, H.; Yamashiro, T.; Otsuki, K.; Ito, T. Development of Reverse Transcription-Loop-Mediated Isothermal Amplification (RT-LAMP) Assay for Detection of Avian Influenza Viruses in Field Specimens. J. Vet. Med. Sci. 2010, 72, 519–523.
- Halami, M.Y.; Dorrestein, G.M.; Couteel, P.; Heckel, G.; Muller, H.; Johne, R. Whole-genome characterization of a novel polyomavirus detected in fatally diseased canary birds. J. Gen. Virol. 2010, 91, 3016–3022.
- Johne, R.; Buck, C.; Allander, T.; Atwood, W.J.; Garcea, R.L.; Imperiale, M.J.; Major, E.O.; Ramqvist, T.; Norkin, L.C. Taxonomical developments in the family Polyomaviridae. Arch. Virol. 2011, 156, 1627–1634.
- Varsani, A.; Porzig, E.L.; Jennings, S.; Kraberger, S.; Farkas, K.; Julian, L.; Massaro, M.; Ballard, G.; Ainley, D.G. Identification of an avian polyomavirus associated with Adélie penguins (Pygoscelis adeliae). J. Gen. Virol. 2015, 96, 851–857.
- Woźniakowski, G.; Tarasiuk, K. Visual detection of goose haemorrhagic polyomavirus in geese and ducks by loop-mediated isothermal amplification. Avian Pathol. 2015, 44, 311–318.
- Zanon, Z.; Najihah, N.; Abu, J.; Mariatulqabtiah, A.R. Prevalence of avian polyomavirus in psittacine birds in the Klang Valley. Pertanika J. Trop. Agric. Sci. 2018, 41, 917–924.
- Johne, R.; Muller, H. Polyomaviruses of Birds: Etiologic Agents of Inflammatory Diseases in a Tumor Virus Family. J. Virol. 2007, 81, 11554–11559.
- Padzil, F.; Mariatulqabtiah, A.; Abu, J. Avian polyomavirus: A recent update. J. Vet. Malays. 2017, 29, 9–13.
- Gaweł, A.; Woźniakowski, G.; Samorek-Salamonowicz, E.; Kozdruń, W.; Bobrek, K.; Bobusia, K.; Nowak, M. Hemorrhagic Nephritis and Enteritis in a Goose Flock in Poland—Disease Course Analysis and Characterization of Etiologic Agent. Avian Dis. 2014, 58, 518–522.
- Park, M.-J.; Kim, H.-R.; Chae, H.-G.; Lim, D.-R.; Kwon, O.-D.; Cho, K.-H.; Park, C.-K. Development of a colorimetric loop-mediated isothermal amplification assay for rapid and specific detection of Aves polyomavirus 1 from psittacine birds. J. Virol. Methods 2019, 273, 113687.
- Parrish, C.R. Papillomaviridae and Polyomaviridae. In Fenner’s Veterinary Virology, 4th ed.; Maclachlan, N.J., Dubovi, E.J., Eds.; Elsevier/AP: Cambridge, MA, USA, 2011; pp. 213–223. ISBN 978-0-1237-5159-1.
- Bert, E.; Tomassone, L.; Peccati, C.; Navarrete, M.G.; Sola, S.C. Detection of Beak and Feather Disease Virus (BFDV) and Avian Polyomavirus (APV) DNA in Psittacine Birds in Italy. J. Vet. Med. Ser. B 2005, 52, 64–68.
- Dolz, G.; Sheleby-Elías, J.; Romero-Zuñiga, J.J.; Vargas-Leitón, B.; Gutiérrez-Espeleta, G.; Madriz-Ordeñana, K. Prevalence of Psittacine Beak and Feather Disease Virus and Avian Polyomavirus in Captivity Psittacines from Costa Rica. Open J. Vet. Med. 2013, 3, 240–245.
- Hsu, C.-M.; Ko, C.-Y.; Tsai, H.-J. Detection and Sequence Analysis of Avian Polyomavirus and Psittacine Beak and Feather Disease Virus from Psittacine Birds in Taiwan. Avian Dis. 2006, 50, 348–353.
- Kuo, Y.H.; Tsai, S.S.; Liu, H.J.; Chuang, K.P. Development of a loop-mediated isothermal amplification method for rapid detection of beak and feather disease virus in parrots. Arch. Clin. Microbiol. 2015, 7, 1–8.
- Woźniakowski, G.; Kozdruń, W.; Samorek-Salamonowicz, E. Loop-mediated isothermal amplification for the detection of goose circovirus. Virol. J. 2012, 9, 110.
- Liu, H.; Li, L.X.; Sun, W.C.; Shi, N.; Sun, X.T.; Jin, N.Y.; Si, X.K. Molecular survey of duck circovirus infection in poultry in southern and southwestern China during 2018 and 2019. BMC Vet. Res. 2020, 16, 80.
- Stenzel, T.; Dziewulska, D.; Muhire, B.M.; Hartnady, P.; Kraberger, S.; Martin, D.P.; Varsani, A. Recombinant Goose Circoviruses Circulating in Domesticated and Wild Geese in Poland. Viruses 2018, 10, 107.
- Tsai, S.S.; Chang, Y.L.; Huang, Y.L.; Liu, H.J.; Ke, G.M.; Chiou, C.J.; Hsieh, Y.C.; Chang, T.C.; Cheng, L.T.; Chuang, K.P. Development of a loop-mediated isothermal amplification method for rapid detection of pigeon circovirus. Arch. Virol. 2014, 159, 921–926.
- Padzil, M.F.M.; Halim, N.S.A.; Najihah, N.; Najian, A.B.N.; Abu, J.; Isa, N.M.; Lau, H.Y.; Mariatulqabtiah, A.R. Evaluation of beak and feather disease virus, avian polyomavirus and avian papillomavirus of captives psittacine birds in Seri Kembangan, Selangor, Malaysia. Malays. J. Microbiol. 2021, 17, 338–344.
- Harkins, G.W.; Martin, D.P.; Christoffels, A.; Varsani, A. Towards inferring the global movement of beak and feather disease virus. Virology 2014, 450–451, 24–33.
- Chae, H.-G.; Lim, D.-R.; Kim, H.-R.; Park, M.-J.; Park, C.-K. An advanced loop-mediated isothermal amplification assay for the rapid detection of beak and feather disease virus in psittacine birds. J. Virol. Methods 2020, 277, 113819.
- Fatoba, A.J.; Adeleke, M. Chicken anemia virus: A deadly pathogen of poultry. Acta Virol. 2019, 63, 19–25.
- Santen, V.L. MSD Veterinary Manual. Available online: https://www.msdvetmanual.com/poultry/chicken-anemia-virus-infection/chicken-anemia-virus-infection (accessed on 15 June 2021).
- Schat, K.A. Chicken Anemia Virus. In TT Viruses. Current Topics in Microbiology and Immunology; de Villiers, E.M., Hausen, H., Eds.; Springer: Berlin, Germany, 2009; Volume 331, pp. 151–183. ISBN 978-3-540-70972-5.
- Huang, C.; Lai, G.; Lee, M.; Lin, W.; Lien, Y.; Hsueh, S.; Kao, J.; Chang, W.; Lu, T.; Chen, H. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of chicken anaemia virus. J. Appl. Microbiol. 2010, 108, 917–924.
- Song, H.; Bae, Y.; Park, S.; Kwon, H.; Lee, H.; Joh, S. Loop-mediated isothermal amplification assay for detection of four immunosuppressive viruses in chicken. J. Virol. Methods 2018, 256, 6–11.
- Woźniakowski, G.; Samorek-Salamonowicz, E. Direct detection of Marek’s disease virus in poultry dust by loop-mediated isothermal amplification. Arch. Virol. 2014, 159, 3083–3087.
- Dunn, J.R.; Gimeno, I. Current Status of Marek’s Disease in the United States and Worldwide Based on a Questionnaire Survey. Avian Dis. 2013, 57, 483–490.
- Bertzbach, L.D.; Conradie, A.M.; You, Y.; Kaufer, B.B. Latest Insights into Marek’s Disease Virus Pathogenesis and Tumorigenesis. Cancers 2020, 12, 647.
- Wei, X.; Shi, X.; Zhao, Y.; Zhang, J.; Wang, M.; Liu, C.; Cui, H.; Hu, S.; Quan, Y.; Chen, H.; et al. Development of a rapid and specific loop-mediated isothermal amplification detection method that targets Marek’s disease virus meq gene. J. Virol. Methods 2012, 183, 196–200.
- Woźniakowski, G.; Samorek-Salamonowicz, E.; Kozdruń, W. Rapid Detection of Marek’s Disease Virus in Feather Follicles by Loop-Mediated Amplification. Avian Dis. 2011, 55, 462–467.
- Wagari, A. A Review on Infectious Bursal Disease in Poultry. Health Econ. Outcome Res. Open Access 2021, 7, 18–23.
- Dey, S.; Pathak, D.C.; Ramamurthy, N.; Maity, H.K.; Chellappa, M.M. Infectious bursal disease virus in chickens: Prevalence, impact, and management strategies. Vet. Med. Res. Rep. 2019, 10, 85–97.
- Wang, Y.; Kang, Z.; Gao, Y.; Qin, L.; Chen, L.; Wang, Q.; Li, J.; Gao, H.; Qi, X.; Lin, H.; et al. Development of loop-mediated isothermal amplification for rapid detection of avian leukosis virus subgroup A. J. Virol. Methods 2011, 173, 31–36.
- Zhang, X.; Liao, M.; Jiao, P.; Luo, K.; Zhang, H.; Ren, T.; Zhang, G.; Xu, C.; Xin, C.; Cao, W. Development of a Loop-Mediated Isothermal Amplification Assay for Rapid Detection of Subgroup J Avian Leukosis Virus. J. Clin. Microbiol. 2010, 48, 2116–2121.
- Chen, H.-T.; Zhang, J.; Ma, Y.-P.; Ma, L.-N.; Ding, Y.-Z.; Liu, X.-T.; Cai, X.-P.; Zhang, Y.-G.; Liu, Y.-S. Reverse transcription loop-mediated isothermal amplification for the rapid detection of infectious bronchitis virus in infected chicken tissues. Mol. Cell. Probes 2010, 24, 104–106.
- El-Tholoth, M.; Mauk, M.G.; Anis, E.; Bau, H.H. A closed-tube, single-step, real time, reverse transcription-loop-mediated isothermal amplification assay for infectious bronchitis virus detection in chickens. J. Virol. Methods 2020, 284, 113940.
- Wu, X.; Song, Z.; Zhai, X.; Zuo, L.; Mei, X.; Xiang, R.; Kang, Z.; Zhou, L.; Wang, H. Simultaneous and visual detection of infectious bronchitis virus and Newcastle disease virus by multiple LAMP and lateral flow dipstick. Poult. Sci. 2019, 98, 5401–5411.
- Li, C.; Chen, Z.; Meng, C.; Liu, G. Rapid detection of duck hepatitis A virus genotype C using reverse transcription loop-mediated isothermal amplification. J. Virol. Methods 2014, 196, 193–198.
- Song, C.; Wan, H.; Yu, S.; Han, X.; Qiu, X.; Hu, Q.; Tan, L.; Ding, C. Rapid detection of duck hepatitis virus type-1 by reverse transcription loop-mediated isothermal amplification. J. Virol. Methods 2012, 182, 76–81.
- Tang, Y.; Yeh, Y.-T.; Chen, H.; Yu, C.; Gao, X.; Diao, Y. Comparison of four molecular assays for the detection of Tembusu virus. Avian Pathol. 2015, 44, 379–385.
- Yan, L.; Peng, S.; Yan, P.; Zhou, J.; Teng, Q.; Li, G.; Li, X.; Li, Z. Comparison of real-time reverse transcription loop-mediated isothermal amplification and real-time reverse transcription polymerase chain reaction for duck Tembusu virus. J. Virol. Methods 2012, 182, 50–55.
- Tunterak, W.; Prakairungnamthip, D.; Ninvilai, P.; Tiawsirisup, S.; Oraveerakul, K.; Sasipreeyajan, J.; Amonsin, A.; Thontiravong, A. Patterns of duck Tembusu virus infection in ducks, Thailand: A serological study. Poult. Sci. 2021, 100, 537–542.
- Li, X.; Shi, Y.; Liu, Q.; Wang, Y.; Li, G.; Teng, Q.; Zhang, Y.; Liu, S.; Li, Z. Airborne Transmission of a Novel Tembusu Virus in Ducks. J. Clin. Microbiol. 2015, 53, 2734–2736.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
14 Feb 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




