
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Chase Melick | + 3645 word(s) | 3645 | 2020-08-31 10:44:27 | | | |
| 2 | Rita Xu | -2338 word(s) | 1307 | 2020-09-07 04:19:18 | | | | |
| 3 | Rita Xu | Meta information modification | 1307 | 2020-10-26 09:20:26 | | |
Video Upload Options
The entry outlines the discovery of mTOR and describes mTOR complex 1 (mTORC1) and mTORC2.
1. Introduction
In 1964, a scientific expedition ventured to Rapa Nui (also known as Easter Island) to collect soil and plants samples [1][2][3]. These samples were brought back to Canada, and rapamycin was isolated from the bacterium Streptomyces hygroscopicus in 1972. Initially, rapamycin was characterized as an antifungal agent, and further studies identified rapamycin to be an immunosuppressant. The ability of rapamycin to inhibit cell growth was discovered later. Experiments demonstrated that rapamycin formed a complex with peptidyl-prolyl cis-trans isomerase FK506-binding protein 12 (FKBP12) [4]. Through genetic screens, the target of rapamycin (TOR) was first discovered in yeast, where mutations in TOR were resistant to rapamycin [5][6][7]. Biochemical experiments in mammalian cells revealed that the rapamycin-FKBP12 complex specifically targets and inhibits the mammalian target of rapamycin (mTOR) [8][9][10]. Through affinity purification, the FKBP12-rapamycin complex was shown to bind a large molecular weight protein called mTOR (also referred to FRAP, RAFT1). Currently, rapamycin and rapamycin analogs (rapalogs) are commonly used as cancer and transplant therapeutics. Decades later, the precise mechanism of how mTOR is regulated is still being elucidated. mTOR coordinates multiple physiological processes through downstream signaling networks.
2. mTOR
mTOR is an evolutionarily conserved Ser/Thr protein kinase that is classified in the phosphatidylinositide 3 kinase (PI3K)-related kinase family within the human phylogenetic kinome tree. mTOR functions as the catalytic subunit of two distinct complexes, referred to as mTORC1 and mTORC2. Rapamycin and rapalogs inhibit mTORC1 activity allosterically, while mTORC2 demonstrates short-term rapamycin insensitivity [11][12][13]. The rapamycin-FKBP12 complex binds to the FKBP12-rapamycin-binding (FRB) domain on mTOR reducing availability of the catalytic cleft, resulting in some substrates unable to access the active site. Prolonged treatment of rapamycin is thought to inhibit mTORC2 through the sequestration of mTOR in some cell types [14][15]. ATP-competitive inhibitors like Torin1 have also been developed, which directly target the catalytic site and inhibit the kinase activity of mTOR [16].
3. mTORC1
mTORC1 consists of three main core components: mTOR, regulatory protein associated with mTOR (Raptor) and mammalian lethal with Sec13 protein 8 (mLST8, also referred to as GβL) (Figure 1, Left) [17][18][19]. Raptor acts as a substrate recognizing subunit that facilitates mTOR phosphorylation through the TOR signaling (TOS) motif found in some mTORC1 substrates [20][21]. Mutations in the TOS motif were shown to render mTORC1 downstream targets, such as the phosphorylation of p70 ribosomal S6 kinase 1 (S6K1) and eIF4E-binding protein 1 (4EBP1, also known as PHAS-1), insensitive to amino acid changes [22]. mLST8 is a positive regulator of mTORC1, stabilizing the association between Raptor and mTOR, and stimulating mTOR kinase activity [19]. mTORC1 contains two additional negative regulators, Proline-rich Akt substrate 40 kDa (PRAS40) [23][24][25] and DEP-domain-containing mTOR-interacting protein (DEPTOR) [26]. PRAS40 acts as a direct inhibitor of substrate binding through the interaction with Raptor, repressing mTORC1 activity [24]. PRAS40 phosphorylation by mTORC1 relieves the negative regulation, increasing mTORC1 signaling [27]. The postsynaptic density 95, discs large, zonula occludens-1 (PDZ) domain of DEPTOR directly interacts with mTOR to inhibit activity [26]. Additionally, mTOR has been shown to promote its own activity via the E3 ubiquitin ligase Skp1, Cullin1, F-box (SCF) adaptor, βTrCP, mediated degradation of DEPTOR [28][29][30].
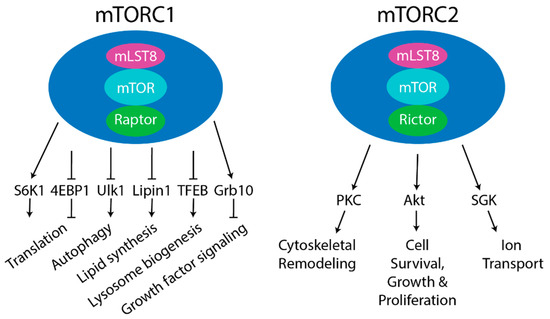
Figure 1. Components of mTOR complex 1 (mTORC1) and mTORC2. Left- Core components of mTORC1 are mammalian target of rapamycin (mTOR) (kinase), Raptor (substrate recognizing component), and mLST8 (positive regulator). Other reported mTORC1 components are PRAS40 (negative regulator) and DEP-domain-containing mTOR-interacting protein (DEPTOR) (negative regulator). Five main downstream pathways are shown. The phosphorylation of S6 kinase 1 (S6K1) and 4EBP1 by mTORC1 regulates protein translation. The phosphorylation of ULK1 by mTORC1 regulates autophagy. mTORC1 also regulates lipid synthesis by phosphorylating S6K1 or Lipin1 to control SREBP, lysosome biogenesis by phosphorylating TFEB, and growth factor signaling by phosphorylating Grb10. Right- Core components of mTORC2 are mTOR (kinase), Rictor (substrate recognizing component), and mLST8 (positive regulator). Other complex components include mSin1 (positive regulator), Protor1/2 (positive regulator), and DEPTOR (negative regulator). mTORC2 regulated processes include cytoskeletal remodeling by phosphorylating PKC; cell survival, growth, and proliferation by phosphorylating Akt; and ion transport by phosphorylating SGK.
When localized to the lysosome, mTORC1 directly interacts with and is activated by the small GTPase Ras homolog enriched in brain (Rheb) [24][31]. However, some mTORC1 mediated process, such as protein translation, presumably occur in the cytoplasm [32]. Additionally, mTORC1 has been observed in other subcellular locations such as the mitochondria [33], stress granules [34], and at the plasma membrane [35]. mTORC1 components have also been reported at multiple locations within the cell [36]. For example, mTOR and Raptor were detected in the nucleus [37]. A more complete discussion of this topic has been reviewed previously [36].
mTORC1 regulates a multitude of cellular processes, such as protein translation, autophagy, lysosome biogenesis, lipid synthesis, and growth factor signaling [38]. mTORC1 regulates translation via the phosphorylation of S6K1 at Thr 389 to activate S6K1 [17]. S6K1 then proceeds to promote translation initiation through the subsequent phosphorylation of factors such as eukaryotic translation initiation factor 4B (eIF4B) [39]. Additionally, mTORC1 phosphorylates 4EBP1 at multiple sites (Thr 37, Thr 46, Ser 65, Thr 70) to promote translation [40]. Once 4EBP1 is phosphorylated it dissociates from eIF4E, which allows the recruitment of the other translation initiation proteins eIF4G and eIF4A [41]. mTORC1 disrupts Unc-51 like autophagy activating kinase 1 (ULK1) interaction with 5′AMP-activated protein kinase (AMPK) through the phosphorylation of Ser 757 (equivalent to Ser 758 in human) on ULK1, to regulate autophagy [42]. Sterol-responsive element-binding protein (SREBP) promotes de novo lipid synthesis [43]. mTORC1 positively regulates SREBP through the phosphorylation and activation of S6K1 or through the multiple site phosphorylation and inhibition of Lipin1, another mTORC1 substrate [43][44][45]. mTORC1 negatively regulates transcription factor EB (TFEB), which promotes genes for lysosomal biogenesis and autophagy machinery at Ser 142 and Ser 211, preventing TFEB nuclear translocation [46][47][48]. Phosphorylation of growth factor receptor-bound protein 10 (Grb10) by mTORC1 at Ser 501 and Ser 503 negatively regulates growth factor signaling through IGF-1 receptor [49][50]. A more comprehensive review of mTORC1 substrates and downstream signaling pathways controlled by mTORC1 is elsewhere [38][39][51].
4. mTORC2
Similar to mTORC1, mTORC2 consists mTOR and mLST8. However, mTORC2 contains rapamycin insensitive companion of mTOR (Rictor) as the substrate recognizing component (Figure 1, Right) [12][13]. Additionally, mTORC2 is comprised of the negative regulator DEPTOR. mTORC2 contains mammalian stress-activated MAPK-interacting protein 1 (mSin1) [52][53][54], which is necessary for the assembly of mTORC2 on the plasma membrane [55]. Activation of mTORC2 depends on the pleckstrin homology (PH) domain of mSin1 that binds to phosphatidylinositol 3,4,5-triphosphate (PtdIns(3,4,5)P3, also referred to as PIP3) at the plasma membrane [56]. Lastly, mTORC2 consists of protein observed with Rictor 1/2 (Protor1/2, also known as PRR5) [57].
mTORC2 has been observed in multiple locations throughout the cell. Using a reporter of endogenous mTORC2 activity, a study showed mTORC2 associates with the plasma membrane, mitochondria, and on endosomal vesicles [58]. mTORC2 has also been reported to localize to the endoplasmic reticulum (ER) and ER associated membranes, such as mitochondria-associated ER membranes (MAMs) [36]. mTORC2 can regulate MAM integrity through the mTORC2 substrate Rac-α Ser/Thr-protein kinase (Akt, also known as PKB) [59]. Lastly, evidence showed mTORC2 may shuttle to the nucleus, however the function of mTORC2 in the nucleus remains unknown [37].
mTORC2 regulates physiological processes through the phosphorylation and activation of downstream substrates like the protein kinase A, G and C (AGC) family. Protein kinase C α (PKCα) at Ser 657 was the first identified substrate of mTORC2 [13]. Other PKC family members have been shown to be phosphorylated and activated by mTORC2, including PKCδ, PKCξ (Thr 560), PKCγ, and PKCε to regulate cytoskeletal remodeling and cell migration [60][61][62]. mTORC2 phosphorylates Akt on Ser 473 to promote cell survival, proliferation and growth [63]. Akt mediates these processes through the subsequent phosphorylation of substrates such as Forkhead box O1/3 (FoxO1/3a) at Thr 32 and Ser 253, glycogen synthase kinase 3 β (GSK3-β) at Ser 9, and tuberous sclerosis complex 2 (TSC2) at Ser 939 and Thr 1462 [54][64][65][66]. Lastly, through the phosphorylation of serum/glucocorticoid-regulated kinase 1 (SGK1) at Ser 422, mTORC2 controls processes like ion transport and cell growth [67]. mTORC2 will not be discussed further in this review, and mTORC2 has been reviewed elsewhere [38][51][68].
References
- Vézina, C.; Kudelski, A.; Sehgal, S.N. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 1975, 28, 721–726, doi:10.7164/antibiotics.28.721.
- Eng, C.P.; Sehgal, S.N.; Vézina, C. Activity of rapamycin (AY-22,989) against transplanted tumors. J. Antibiot. 1984, 37, 1231–1237, doi:10.7164/antibiotics.37.1231.
- Martel, R.R.; Klicius, J.; Galet, S. Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can. J. Physiol. Pharmacol. 1977, 55, 48–51, doi:10.1139/y77-007.
- Chung, J.; Kuo, C.J.; Crabtree, G.R.; Blenis, J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 1992, 69, 1227–1236, doi:10.1016/0092-8674(92)90643-q.
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909, doi:10.1126/science.1715094.
- Kunz, J.; Henriquez, R.; Schneider, U.; Deuter-Reinhard, M.; Movva, N.; Hall, M.N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 1993, 73, 585–596, doi:10.1016/0092-8674(93)90144-f.
- Cafferkey, R.; Young, P.R.; McLaughlin, M.M.; Bergsma, D.J.; Koltin, Y.; Sathe, G.M.; Faucette, L.; Eng, W.K.; Johnson, R.K.; Livi, G.P. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 1993, 13, 6012–6023, doi:10.1128/mcb.13.10.6012.
- Brown, E.J.; Albers, M.W.; Shin, T.B.; Ichikawa, K.; Keith, C.T.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by G1-arresting rapamycin–receptor complex. Nature 1994, 369, 756–758, doi:10.1038/369756a0.
- Sabatini, D.M.; Erdjument-Bromage, H.; Lui, M.; Tempst, P.; Snyder, S.H. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994, 78, 35–43, doi:10.1016/0092-8674(94)90570-3.
- Sabers, C.J.; Martin, M.M.; Brunn, G.J.; Williams, J.M.; Dumont, F.J.; Wiederrecht, G.; Abraham, R.T. Isolation of a Protein Target of the FKBP12-Rapamycin Complex in Mammalian Cells. J. Biol. Chem. 1995, 270, 815–822, doi:10.1074/jbc.270.2.815.
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468, doi:10.1016/s1097-2765(02)00636-6.
- Jacinto, E.; Loewith, R.; Schmidt, A.; Lin, S.; Rüegg, M.A.; Hall, A.; Hall, M.N. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004, 6, 1122–1128, doi:10.1038/ncb1183.
- Sarbassov, D.D.; Ali, S.M.; Kim, D.-H.; Guertin, D.A.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. Rictor, a Novel Binding Partner of mTOR, Defines a Rapamycin-Insensitive and Raptor-Independent Pathway that Regulates the Cytoskeleton. Curr. Biol. 2004, 14, 1296–1302, doi:10.1016/j.cub.2004.06.054.
- Sarbassov, D.D.; Ali, S.M.; Sengupta, S.; Sheen, J.-H.; Hsu, P.P.; Bagley, A.F.; Markhard, A.L.; Sabatini, D.M. Prolonged Rapamycin Treatment Inhibits mTORC2 Assembly and Akt/PKB. Mol. Cell 2006, 22, 159–168, doi:10.1016/j.molcel.2006.03.029.
- Phung, T.L.; Ziv, K.; Dabydeen, D.; Eyiah-Mensah, G.; Riveros, M.; Perruzzi, C.; Sun, J.; Monahan-Earley, R.A.; Shiojima, I.; Nagy, J.A.; et al. Pathological angiogenesis is induced by sustained Akt signaling and inhibited by rapamycin. Cancer Cell 2006, 10, 159–170, doi:10.1016/j.ccr.2006.07.003.
- Thoreen, C.C.; Kang, S.A.; Chang, J.W.; Liu, Q.; Zhang, J.; Gao, Y.; Reichling, L.J.; Sim, T.; Sabatini, D.M.; Gray, N.S. An ATP-competitive Mammalian Target of Rapamycin Inhibitor Reveals Rapamycin-resistant Functions of mTORC1. J. Biol. Chem. 2009, 284, 8023–8032, doi:10.1074/jbc.m900301200.
- Kim, D.-H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR Interacts with Raptor to Form a Nutrient-Sensitive Complex that Signals to the Cell Growth Machinery. Cell 2002, 110, 163–175, doi:10.1016/s0092-8674(02)00808-5.
- Hara, K.; Maruki, Y.; Long, X.; Yoshino, K.-I.; Oshiro, N.; Hidayat, S.; Tokunaga, C.; Avruch, J.; Yonezawa, K. Raptor, a Binding Partner of Target of Rapamycin (TOR), Mediates TOR Action. Cell 2002, 110, 177–189, doi:10.1016/s0092-8674(02)00833-4.
- Kim, D.-H.; Sarbassov, D.D.; Ali, S.M.; Latek, R.R.; Guntur, K.V.P.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 2003, 11, 895–904.
- Nojima, H.; Tokunaga, C.; Eguchi, S.; Oshiro, N.; Hidayat, S.; Yoshino, K.-I.; Hara, K.; Tanaka, N.; Avruch, J.; Yonezawa, K. The Mammalian Target of Rapamycin (mTOR) Partner, Raptor, Binds the mTOR Substrates p70 S6 Kinase and 4E-BP1 through Their TOR Signaling (TOS) Motif. J. Biol. Chem. 2003, 278, 15461–15464, doi:10.1074/jbc.c200665200.
- Schalm, S.S.; Fingar, D.C.; Sabatini, D.M.; Blenis, J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 2003, 13, 797–806, doi:10.1016/s0960-9822(03)00329-4.
- Schalm, S.S.; Blenis, J. Identification of a Conserved Motif Required for mTOR Signaling. Curr. Biol. 2002, 12, 632–639, doi:10.1016/s0960-9822(02)00762-5.
- Haar, E.V.; Lee, S.-I.; Bandhakavi, S.; Griffin, T.J.; Kim, D.-H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007, 9, 316–323, doi:10.1038/ncb1547.
- Sancak, Y.; Thoreen, C.C.; Peterson, T.R.; Lindquist, R.A.; Kang, S.A.; Spooner, E.; Carr, S.A.; Sabatini, D.M. PRAS40 Is an Insulin-Regulated Inhibitor of the mTORC1 Protein Kinase. Mol. Cell 2007, 25, 903–915, doi:10.1016/j.molcel.2007.03.003.
- Wang, L.; Harris, T.E.; Roth, R.A.; Lawrence, J.C. PRAS40 Regulates mTORC1 Kinase Activity by Functioning as a Direct Inhibitor of Substrate Binding. J. Biol. Chem. 2007, 282, 20036–20044, doi:10.1074/jbc.m702376200.
- Peterson, T.R.; Laplante, M.; Thoreen, C.C.; Sancak, Y.; Kang, S.A.; Kuehl, W.M.; Gray, N.S.; Sabatini, D.M. DEPTOR Is an mTOR Inhibitor Frequently Overexpressed in Multiple Myeloma Cells and Required for Their Survival. Cell 2009, 137, 873–886, doi:10.1016/j.cell.2009.03.046.
- Wang, L.; Harris, T.E.; Lawrence, J.C. Regulation of Proline-rich Akt Substrate of 40 kDa (PRAS40) Function by Mammalian Target of Rapamycin Complex 1 (mTORC1)-mediated Phosphorylation. J. Biol. Chem. 2008, 283, 15619–15627, doi:10.1074/jbc.m800723200.
- Duan, S.; Skaar, J.R.; Kuchay, S.; Toschi, A.; Kanarek, N.; Ben-Neriah, Y.; Pagano, M. mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR. Mol. Cell 2011, 44, 317–324.
- Gao, D.; Inuzuka, H.; Tan, M.K.; Fukushima, H.; Locasale, J.W.; Liu, P.; Wan, L.; Zhai, B.; Chin, Y.R.; Shaik, S.; et al. mTOR drives its own activation via SCF(betaTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol. Cell 2011, 44, 290–303.
- Zhao, Y.; Xiong, X.; Sun, Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(betaTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol. Cell 2011, 44, 304–316.
- Long, X.; Lin, Y.; Ortiz-Vega, S.; Yonezawa, K.; Avruch, J. Rheb Binds and Regulates the mTOR Kinase. Curr. Biol. 2005, 15, 702–713, doi:10.1016/j.cub.2005.02.053.
- Holz, M.K.; Ballif, B.A.; Gygi, S.P.; Blenis, J. mTOR and S6K1 Mediate Assembly of the Translation Preinitiation Complex through Dynamic Protein Interchange and Ordered Phosphorylation Events. Cell 2005, 123, 569–580, doi:10.1016/j.cell.2005.10.024.
- Paglin, S. Rapamycin-Sensitive Pathway Regulates Mitochondrial Membrane Potential, Autophagy, and Survival in Irradiated MCF-7 Cells. Cancer Res. 2005, 65, 11061–11070, doi:10.1158/0008-5472.can-05-1083.
- Thedieck, K.; Holzwarth, B.; Prentzell, M.T.; Boehlke, C.; Klasener, K.; Ruf, S.; Sonntag, A.G.; Maerz, L.; Grellscheid, S.N.; Kremmer, E.; et al. lInhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cels. Cell 2013, 154, 859–874.
- Bridges, D.; Ma, J.-T.; Park, S.; Inoki, K.; Weisman, L.S.; Saltiel, A. Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol. Biol. Cell 2012, 23, 2955–2962, doi:10.1091/mbc.e11-12-1034.
- Betz, C.; Hall, M.N. Where is mTOR and what is it doing there? J. Cell Biol. 2013, 203, 563–574, doi:10.1083/jcb.201306041.
- Rosner, M.; Hengstschläger, M. Cytoplasmic and nuclear distribution of the protein complexes mTORC1 and mTORC2: Rapamycin triggers dephosphorylation and delocalization of the mTORC2 components rictor and sin1. Hum. Mol. Genet. 2008, 17, 2934–2948, doi:10.1093/hmg/ddn192.
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 169, 361–371.
- Ma, X.M.; Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009, 10, 307–318, doi:10.1038/nrm2672.
- Gingras, A.-C.; Raught, B.; Gygi, S.P.; Niedźwiecka-Kornaś, A.; Miron, M.; Burley, S.K.; Polakiewicz, R.D.; Wyslouch-Cieszynska, A.; Aebersold, R.; Sonenberg, N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genome Res. 2001, 15, 2852–2864.
- Gingras, A.-C.; Raught, B.; Sonenberg, N. eIF4 Initiation Factors: Effectors of mRNA Recruitment to Ribosomes and Regulators of Translation. Annu. Rev. Biochem. 1999, 68, 913–963, doi:10.1146/annurev.biochem.68.1.913.
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141, doi:10.1038/ncb2152.
- Porstmann, T.; Santos, C.R.; Griffiths, B.; Cully, M.; Wu, M.; Leevers, S.; Griffiths, J.R.; Chung, Y.-L.; Schulze, A. SREBP Activity Is Regulated by mTORC1 and Contributes to Akt-Dependent Cell Growth. Cell Metab. 2008, 8, 224–236, doi:10.1016/j.cmet.2008.07.007.
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR Complex 1 Regulates Lipin 1 Localization to Control the SREBP Pathway. Cell 2011, 146, 408–420, doi:10.1016/j.cell.2011.06.034.
- Düvel, K.; Yecies, J.L.; Menon, S.; Raman, P.; Lipovsky, A.I.; Souza, A.L.; Triantafellow, E.; Ma, Q.; Gorski, R.; Cleaver, S.; et al. Activation of a Metabolic Gene Regulatory Network Downstream of mTOR Complex 1. Mol. Cell 2010, 39, 171–183, doi:10.1016/j.molcel.2010.06.022.
- Settembre, C.; Zoncu, R.; Medina, D.L.; Vetrini, F.; Erdin, S.; Erdin, S.; Huynh, T.; Ferron, M.; Karsenty, G.; Vellard, M.C.; et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012, 31, 1095–1108, doi:10.1038/emboj.2012.32.
- Martina, J.A.; Chen, Y.; Gucek, M.; Puertollano, R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012, 8, 903–914, doi:10.4161/auto.19653.
- Roczniak-Ferguson, A.; Petit, C.S.; Froehlich, F.; Qian, S.; Ky, J.; Angarola, B.; Walther, T.C.; Ferguson, S.M. The Transcription Factor TFEB Links mTORC1 Signaling to Transcriptional Control of Lysosome Homeostasis. Sci. Signal. 2012, 5, ra42, doi:10.1126/scisignal.2002790.
- Hsu, P.P.; Kang, S.A.; Rameseder, J.; Zhang, Y.; Ottina, K.A.; Lim, D.; Peterson, T.R.; Choi, Y.; Gray, N.S.; Yaffe, M.B.; et al. The mTOR-Regulated Phosphoproteome Reveals a Mechanism of mTORC1-Mediated Inhibition of Growth Factor Signaling. Science 2011, 332, 1317–1322, doi:10.1126/science.1199498.
- Yu, Y.; Yoon, S.-O.; Poulogiannis, G.; Yang, Q.; Ma, X.M.; Villén, J.; Kubica, N.; Hoffman, G.R.; Cantley, L.C.; Gygi, S.P.; et al. Phosphoproteomic Analysis Identifies Grb10 as an mTORC1 Substrate That Negatively Regulates Insulin Signaling. Science 2011, 332, 1322–1326, doi:10.1126/science.1199484.
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203, doi:10.1038/s41580-019-0199-y.
- Yang, Q.; Inoki, K.; Ikenoue, T.; Guan, K.-L. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006, 20, 2820–2832, doi:10.1101/gad.1461206.
- Frías, M.A.; Thoreen, C.C.; Jaffe, J.D.; Schroder, W.; Sculley, T.; Carr, S.A.; Sabatini, D.M. mSin1 Is Necessary for Akt/PKB Phosphorylation, and Its Isoforms Define Three Distinct mTORC2s. Curr. Biol. 2006, 16, 1865–1870, doi:10.1016/j.cub.2006.08.001.
- Jacinto, E.; Facchinetti, V.; Liu, D.; Soto, N.; Wei, S.; Jung, S.Y.; Huang, Q.; Qin, J.; Su, B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 2006, 127, 125–137.
- Yuan, H.-X.; Guan, K.-L. The SIN1-PH Domain Connects mTORC2 to PI3K. Cancer Discov. 2015, 5, 1127–1129, doi:10.1158/2159-8290.cd-15-1125.
- Liu, P.; Gan, W.; Chin, Y.R.; Ogura, K.; Guo, J.; Zhang, J.; Wang, B.; Blenis, J.; Cantley, L.C.; Toker, A.; et al. PtdIns(3,4,5)P3-Dependent Activation of the mTORC2 Kinase Complex. Cancer Discov. 2015, 5, 1194–1209, doi:10.1158/2159-8290.cd-15-0460.
- Pearce, L.R.; Huang, X.; Boudeau, J.; Pawłowski, R.; Wullschleger, S.; Deak, M.; Ibrahim, A.F.M.; Gourlay, R.; Magnuson, M.A.; Alessi, D.R. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem. J. 2007, 405, 513–522, doi:10.1042/bj20070540.
- Ebner, M.; Sinkovics, B.; Szczygieł, M.; Ribeiro, D.W.; Yudushkin, I.A. Localization of mTORC2 activity inside cells. J. Cell Biol. 2017, 216, 343–353, doi:10.1083/jcb.201610060.
- Betz, C.; Stracka, D.; Prescianotto-Baschong, C.; Frieden, M.; Demaurex, N.; Hall, M.N. Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc. Natl. Acad. Sci. USA 2013, 110, 12526–12534.
- Gan, X.; Wang, J.; Wang, C.; Sommer, E.; Kozasa, T.; Srinivasula, S.; Alessi, D.; Offermanns, S.; Simon, M.I.; Wu, D. PRR5L degradation promotes mTORC2-mediated PKC-delta phosphorylation and cell migration downstream of Galpha12. Nat. Cell Biol. 2012, 14, 686–696.
- Li, X.; Gao, T. mTORC2 phosphorylates protein kinase Czeta to regulate its stability and activity. EMBO Rep. 2014, 15, 191–198.
- Thomanetz, V.; Angliker, N.; Cloëtta, D.; Lustenberger, R.M.; Schweighauser, M.; Oliveri, F.; Suzuki, N.; Rüegg, M.A. Ablation of the mTORC2 component rictor in brain or Purkinje cells affects size and neuron morphology. J. Cell Biol. 2013, 201, 293–308, doi:10.1083/jcb.201205030.
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and Regulation of Akt/PKB by the Rictor-mTOR Complex. Science 2005, 307, 1098–1101, doi:10.1126/science.1106148.
- Guertin, D.A.; Stevens, D.M.; Thoreen, C.C.; Burds, A.A.; Kalaany, N.Y.; Moffat, J.; Brown, M.; Fitzgerald, K.J.; Sabatini, D.M. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell 2006, 11, 859–871.
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt Promotes Cell Survival by Phosphorylating and Inhibiting a Forkhead Transcription Factor. Cell 1999, 96, 857–868, doi:10.1016/s0092-8674(00)80595-4.
- Manning, B.D.; Tee, A.R.; Logsdon, M.N.; Blenis, J.; Cantley, L.C. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 2002, 10, 151–162, doi:10.1016/s1097-2765(02)00568-3.
- García-Martínez, J.M.; Alessi, D.R. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem. J. 2008, 416, 375–385, doi:10.1042/bj20081668.
- Oh, W.J.; Jacinto, E. mTOR complex 2 signaling and functions. Cell Cycle 2011, 10, 2305–2316, doi:10.4161/cc.10.14.16586.




