Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Marina Schramm | + 5683 word(s) | 5683 | 2022-01-30 09:21:06 | | | |
| 2 | Camila Xu | -1979 word(s) | 3704 | 2022-02-07 09:22:53 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Schramm, M. Cell-Free Protein Synthesis of Unspecific Peroxygenases. Encyclopedia. Available online: https://encyclopedia.pub/entry/19134 (accessed on 08 February 2026).
Schramm M. Cell-Free Protein Synthesis of Unspecific Peroxygenases. Encyclopedia. Available at: https://encyclopedia.pub/entry/19134. Accessed February 08, 2026.
Schramm, Marina. "Cell-Free Protein Synthesis of Unspecific Peroxygenases" Encyclopedia, https://encyclopedia.pub/entry/19134 (accessed February 08, 2026).
Schramm, M. (2022, February 07). Cell-Free Protein Synthesis of Unspecific Peroxygenases. In Encyclopedia. https://encyclopedia.pub/entry/19134
Schramm, Marina. "Cell-Free Protein Synthesis of Unspecific Peroxygenases." Encyclopedia. Web. 07 February, 2022.
Copy Citation
Unspecific peroxygenases (UPOs, EC 1.11.2.1) are fungal biocatalysts that have attracted considerable interest for application in chemical syntheses due to their ability to selectively incorporate peroxide-oxygen into non-activated hydrocarbons. However, the number of available and characterized UPOs is limited, as it is difficult to produce these enzymes in homologous or hetero-logous expression systems.
unspecific peroxygenase
EC 1.11.2.1
monooxygenase
cell-free protein synthesis
in vitro translation
1. Introduction
Over the last two decades, biocatalysis has increasingly developed into a mature and widely applicable technology for chemical synthesis and production [1]. A number of bio-catalytic approaches have been developed for the environmentally friendly and effective synthesis of a range of chiral products, such as pharmaceuticals, agrochemicals, and fine chemicals. However, further development of these biocatalytic processes on an industrial scale will depend crucially on the widespread availability of efficient and robust enzymes [2][3].
Unspecific peroxygenases (EC 1.11.2.1) are enzymes found in numerous fungi and are ideally suited for the synthesis of specialty chemicals. As extracellular enzymes, they are inherently stable and self-sufficient, as evidenced by their ease of activation by hydrogen peroxide and independence of electron transport proteins, electron donors, or cofactors other than heme-stabilizing magnesium ions [4][5]. They have become ‘dream catalysts’ in organic synthesis because of their ability to selectively incorporate peroxidic oxy-gen into non-activated hydrocarbons, causing hydroxylations and epoxidations that are difficult to achieve by chemical methods [6][7][8][9]. Essential for this type of reaction is the heme cofactor that is coordinated proximally by the thiol of a cysteine residue. Due to their P450-like catalytic mechanism and reaction spectrum, UPOs can be used for the conversion of pharmaceuticals and drugs, e.g., of volixibat (N-dealkylation) [10], cyclophosphamide (aliphatic hydroxylation) [11], propranolol (aromatic hydroxylation) [12], testosterone (epoxidation) [13], corticosteroids (side-chain removal by C-C scission) [14] and clopidogrel (sulfoxidation and epoxidation) [15].
A crucial drawback in the application of UPOs (representing a diverse superfamily of proteins) and the main reason why their scope is still very narrow is their low avail-ability, limited to a few representatives [16]. Contrary to their undemanding nature in catalysis, UPOs are demanding when it comes to their expression. To date, there are only a few reports demonstrating the efficient production and purification of UPOs using wild-type fungi [13][14][17][18][19][20]. To circumvent the disadvantages of homologous production, such as low production rates, long production times, or high concentrations of contaminating proteins, various attempts have been made to express UPOs heterologously in established hosts. An overview of recombinantly expressed UPOs is given in the recent review of Kinner and colleagues [21]. Another peroxygenase from Hypoxylon that has been recently produced in Pichia pastoris (syn. Komagataella phaffii) can be added to this list [22].
However, even when using quite different host organisms, such as Saccharomyces cerevisiae, P. pastoris, Aspergillus spp., or Escherichia coli, it has not yet been possible to establish a universal expression system for UPOs. Overall, the number of available UPOs today is less than 20, which is in stark contrast to the number of putative UPO genes in fungal genomes, which is in the range of several thousand [20]. To close the gap between theoretically and practically available UPOs and to gain access to new catalytically promising candidates, a third option (besides homologous and heterologous expression) was chosen in this study: the production of UPOs by cell-free protein synthesis (CFPS).
There are examples of functional heme-containing enzymes that were cell-free expressed using lysates from E. coli: manganese peroxidase [23][24], a P450 monooxygenase [25], cytochrome c oxidase [26], and horseradish peroxidase [27]. This demonstrates the general potential of CFPS for the production of enzymes with such a complex prosthetic group; however, similar results have not yet been published regarding UPOs.
In general, CFPS can be considered a powerful and versatile tool for the rapid discovery of new biocatalyst candidates or improved biocatalyst variants [28]. Optimized systems are also of interest for the preparative production of proteins with yields in the milligram-per-milliliter range. There are CFPS systems based on bacterial, plant, archaeal, fungal, and animal cells. The most relevant and well-studied lysates used for CFPS are from E. coli, Spodoptera frugiperda (insect), S. cerevisiae (yeast), Chinese hamster ovary (CHO), rabbit reticulocyte, wheat germ, and HeLa cells [29]. To date, filamentous fungi just play a minor role in CFPS. In some published work, lysates from Neurospora crassa have been employed to evaluate the potential for the production of cell-free expressed proteins using reporter proteins or for in vitro studies as such, for example, to investigate the impact of codon usage [30][31][32][33]. Regarding the genus Aspergillus, there is only one publication dealing with the use of a lysate from A. nidulans in CFPS [34]. No studies have been published on A. niger in the context of CFPS.
N. crassa and A. niger are filamentous fungi from the phylum Ascomycota, but belong to different classes (Sordariomycetes and Eurotiomycetes, respectively). Based on bioinformatic analyses, both organisms carry putative UPO genes in their genomes [20]. There are good reasons for using filamentous fungi in CFPS, such as their innate competence for post-translational modifications, their ability to secrete proteins, and also their low requirements for the growth medium, which makes the preparation of lysates inexpensive.
2. Lysate Preparation from Neurospora crassa and Aspergillus niger
N. crassa and A. niger were grown in nutrient-rich media for 48 h and 24 h, respectively. Subsequently, the mycelia were harvested by vacuum filtration and yielded fresh weights of 25 to 30 g L−1 for both fungi. For cell disruption, a French press homogenizer was used, which represents a simple and fast method and is routinely used in the preparation of lysates from other fungal organisms such as S. cerevisiae [35]. The lysis was successful as 80 to 90% of the cells were broken, which was microscopically detectable (Figure 1a).
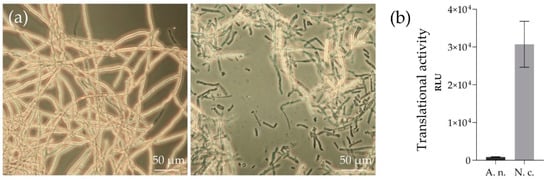
Figure 1. Lysis of fungal cells and testing of translationally active lysates from N. crassa and A. niger. (a) Mycelium of N. crassa prior to (left panel) and after (right panel) lysis with a French press homogenizer. Intact cells appear bright. (b) Cell-free expressed luciferase with lysates from A. niger (A. n.) and N. crassa (N. c.). The 20-µL reactions were incubated at 18 °C for 90 min and tested for luciferase activity by measuring the oxidation of luciferin via luminescence detection. The experiment was done in duplicate. RLU—relative luminescence units.
The lysates were tested for translational activities by using the cytosolic reporter enzyme ‘firefly luciferase’ (Figure 1b). With both lysates, significant amounts of firefly luci-ferase were in vitro translated, albeit in differing quantities. The measured values for N. crassa (30,721 ± 6091 RLU) were 35-times higher than those of A. niger (862 ± 78 RLU).
3. Cell-Free Protein Synthesis of AaeUPO
For our study on the cell-free production of a UPO, we used the upo1 gene from Cyclocybe (Agrocybe) aegerita, which encodes the first reported UPO [17]. The extracellular heme-thiolate enzyme contains an N-terminal signal peptide followed by a propeptide and has a C-terminal intramolecular disulfide bond as well as six potential N-glycosylation sites [36]. For easier detection, the UPO gene was tagged with a C-terminal polyhistidine peptide. To reduce the complexity of the CFPS system, the mRNA of upo1 was synthesized independently from the translation system and provided externally. The CFPS reactions were tested for the formation of functional cell-free UPO with an LC-MS-based propranolol assay since the formation of 5-hydroxypropranolol (5-OHP) from propranolol is characteristic of AaeUPO [12][37]. By using this assay, we could exclude the possibility of measuring false positive activities from potentially interfering endogenous monooxygenases (cytochrome P450 enzymes). Nevertheless, negative controls that contained no mRNA templates were additionally included in all experimental setups.
Cell-free production of AaeUPO was evaluated with lysates from N. crassa and A. niger, respectively. Essentially, the reactions were set up according to Wu et al. with minor modifications [33]. The mRNA carrying the coding sequence for the UPO was applied at 28 nM since higher concentrations had adverse effects on the synthesis of active enzyme (Figure A1). The formation of active cfAaeUPO was observed over a period of 24 h (Figure 2). With both lysates, significant UPO activity could be detected: 10,498 ± 28 mU L−1 using the N. crassa lysate and 783 ± 18 mU L−1 with the A. niger lysate. The different yields of active enzyme correlated with the results for the reporter luciferase (Figure 1b). Compared to the lysate from N. crassa, the yield of active enzyme with the A. niger lysate was 10-times (after 5 h incubation) to 15-times (after 24 h incubation) lower. Interestingly, in contrast to N. crassa, the UPO activity with the A. niger lysate was higher after 5 h of incubation than after 24 h. This decrease in activity could indicate residual proteolytic activity in the lysate, despite the application of protease inhibitors during the preparation process.
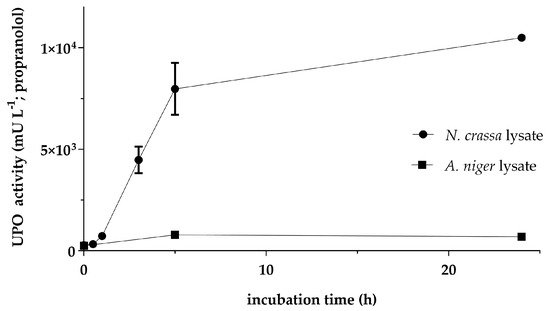
Figure 2. Time-dependent activity of cell-free synthesized AaeUPO with lysates from N. crassa and A. niger. The 20-µL reactions were prepared in separate tubes, incubated at 18 °C for the indicated time, and tested for UPO activity by measuring the formation of 5-OHP from propranolol by LC-MS analysis. Values are means ± standard deviation.
Western blot analyses were done to get information about the localization of the synthesized protein and its relative amount (Figure 3). As a secretory protein, cfAaeUPO was expected to be present in the microsomes, i.e., in ER-derived vesicles. In the case of the A. niger lysate-based CFPS, a band was detected in the microsomal fraction that corresponded to AaeUPO, which has a theoretical molecular weight of 41.5 kDa in its unprocessed form. Overall, the amount of detectable protein was rather low, which explains the relatively low activity. With the N. crassa lysate, two prominent bands could be seen in the whole CFPS reaction mixture. As demonstrated by deglycosylation (Figure 3d), the lower band corresponds to the non-glycosylated (marked with an asterisk) and the upper band to the glycosylated (marked with an arrowhead) form of cfAaeUPO. The larger part of the glycosylated protein was located in the microsomes, while the non-glycosylated UPO was almost exclusively found in the supernatant. Activity measurements of the fractions indicated that the UPO activity stems from the glycosylated form as the ratio of the activities between the fractions correlated with the distribution of synthesized protein (Figure A2). The residual glycosylated cfAaeUPO seen in the supernatant fraction may be the result of an incomplete fractionation process, similarly described in an article by Thoring et al. [38]. They showed that—after centrifugation with 16,000× g—smaller microsomes were still present in the supernatant. The rather high portion of non-glycosylated protein might indicate an insufficient translocation capacity. Nonetheless, the successful expression of active AaeUPO proves that our fungal lysates contain functional microsomes that allow appropriate post-translational modification of secretory proteins.
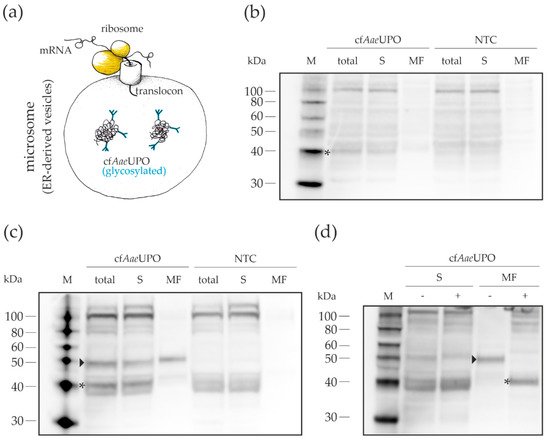
Figure 3. Localization of cell-free AaeUPO synthesized with lysates from A. niger and N. crassa, respectively. CFPS was done in 40-µL reactions, incubated at 18 °C for 5 h and fractionated; 10 µL of the whole CFPS reactions, supernatant, and microsomal fractions, respectively, were subjected to SDS-PAGE and subsequent Western blot analysis using a polyhistidine tag antibody. (a) Schematic representation of the cell-free synthesis of AaeUPO. (b) CFPS samples from A. niger. (c) CFPS samples from N. crassa. (d) Deglycosylation of cfAaeUPO produced with lysate from N. crassa. (−)—without deglycosylation enzyme mix; (+) with deglycosylation enzyme mix; M—protein marker, total—whole CFPS reaction; S—supernatant; MF—microsomal fraction; NTC—no-template control; arrowhead—glycosylated cfAaeUPO; asterisk—non-glycosylated cfAaeUPO.
Unfortunately, it was not possible to use the polyhistidine tag on the cfAaeUPO for purification from the CFPS reaction with sufficient purity, due to the presence of a larger amount of contaminating fungal proteins. Thus, an accurate protein concentration of the synthesized cfAaeUPO could not be determined. In future experiments, the use of more appropriate affinity tags, such as the Strep II tag, should be considered [39].
The good results for the long incubation times with the lysate from N. crassa (Figure 2) were unexpected since, in batch CFPS reactions, the synthesis process usually stops after a few hours. For example, Hodgman and Jewett reported a “long synthesis time of 120 min” for luciferase using a yeast CFPS-batch reaction system, albeit at a slightly higher incubation temperature of 21 °C [40]. The main reasons for the termination of synthesis were seen in a rapid decline in energy-providing components and an accumulation of inhibitory molecules, such as inorganic phosphate [41]. To clarify whether the apparently long synthesis time in our experiments was actually due to better protein synthesis performance of the lysate, we expressed the reporter protein luciferase using CFPS with the lysate of N. crassa and measured the activity over a 24-hour period (Figure 4a). The activity of luciferase increased over the first three hours of CFPS but decreased thereafter, suggesting that the process of protein synthesis was not maintained throughout the incubation period. To confirm that this was also true for CFPS of AaeUPO, samples from the time-course experiment (Figure 2) were subjected to Western blot analysis (Figure 4b). As in the previous Western blot, two prominent bands appeared in the gel. Their intensity and thus the amount of cfAaeUPO produced increased until the third hour of incubation and then remained nearly constant (or decreased slightly) for the next hours. The 24-hour samples appeared to have lower cfAaeUPO protein levels than the samples from the previous time points, although activity was highest after 24 h. It might be that nonfunctional, e.g., misfolded UPO protein was degraded by remaining proteases in the lysate, but this needs to be clarified in future experiments.
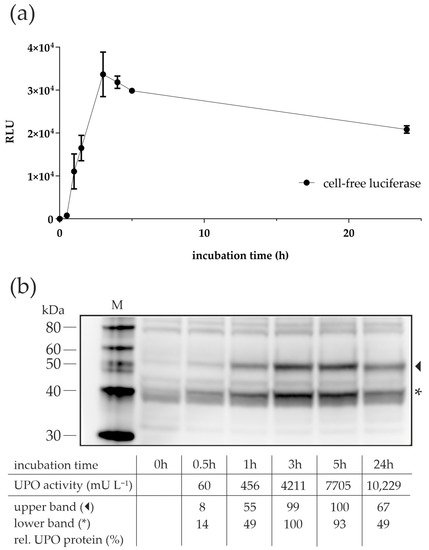
Figure 4. Time-dependent cell-free protein synthesis of luciferase and AaeUPO with N. crassa lysate. (a) Detection of luciferase activity in CFPS-batch reactions at different time points. The 20-µL reactions were prepared in separate tubes, incubated at 18 °C for the indicated time, and tested for luciferase activity by measuring the oxidation of luciferin via luminescence detection. Values are means ± standard deviation. (b) Western blot analysis of CFPS samples from different incubation times. The 20-µL reactions were prepared in separate tubes, incubated at 18 °C for the indicated time, and subjected to SDS-PAGE and, subsequently, Western blot analyses using a polyhistidine tag antibody. The semi-quantitative analyses were done individually for the glycosylated and non-glycosylated proteins. The densitometric values for t0 were subtracted from the values of other samples and the highest values were set to 100%. M—protein marker; arrowhead—glycosylated cfAaeUPO; asterisk—non-glycosylated cfAaeUPO.
Since the capacity to produce protein was limited to 3 h in our setup, an intrinsic maturation period of the UPO protein could be an explanation for the apparently long synthesis time. Such a maturation time was observed for a bacterial P450 monooxygenase that was expressed in a cell-free approach using lysate from E. coli [25]. In this case, the highest enzyme activity was reached 150 min after protein synthesis had stopped. However, since the exact process of heme incorporation and protein folding is still unknown for UPOs, it must be clarified in the future whether a longer maturation time is indeed necessary. Here, the CFPS might prove to be a valuable tool for gaining insight into the genesis of UPOs. It should be relatively simple to change the conditions in CFPS, for example, by adding mRNA for the co-expression of chaperones. The effects on UPO formation would be directly measurable and could be evaluated in a short time.
Table 1 gives a brief comparison of the volume activities and production times of AaeUPOs obtained from homologous, heterologous, or cell-free approaches. Attempts to express wtAaeUPO heterologously in the standard expression hosts S. cerevisiae or P. pastoris with appreciable yields failed [42][43]. Rounds of directed evolution were necessary to increase the production in the two yeasts that do not contain endogenous UPO genes [20][43]. This highlights an advantage of the cell-free approach: the possibility to use translationally active lysates from naturally UPO-coding (and producing) fungi. Thereby, all essential components should be available for the in vitro UPO synthesis. However, it has to be mentioned that the evolved mutant variant rAaeUPO PaDa-I expressed in P. pastoris in a bioreactor in a six-day cultivation period reached much higher expression levels than the homologous or cell-free systems. Nevertheless, cell-free protein production is by far the fastest method to gain significant amounts of functional UPO.
Table 1. Comparison of AaeUPO produced by different approaches.
If the heterologously produced PaDa-I variant of rAaeUPO is included in the comparison of the three expression systems, the yield obtained with CFPS appears to be the lowest. However, in the present study, we have not yet fully exploited the potential of the cell-free approach using fungal lysates. There are general aspects in CFPS that have a high impact on the performance and would need to be optimized, for example, biomass production, lysate preparation, and buffer composition [44][45]. Also, the template design is essential for an optimal translation initiation, which can be triggered by adding elements like IRES [46]. As indicated by the Western blot analyses (Figure 3), the production of active UPO in our setup may be limited by a non-optimal translocation efficiency. Co-expressing components of the secretory apparatus could solve this problem, as this approach was already successful in living cells, for example, for the production of secreted antibodies in CHO cells [47]. Additionally, there is the possibility to change the CFPS reaction from a batch to a continuous process, which would result in an increase in protein yield [48]. Other influential parameters or components are specific for the expression of UPOs. The availability and fit of the heme cofactor are crucial for the functioning of UPOs, therefore the formulation and amount of heme in CFPS reactions will have to be optimized. This could be achieved on different levels: (i) during growth of the fungi, e.g., with the addition of hemin, which worked well in the expression of horseradish peroxidase in P. pastoris [49]; and (ii) by adding heme directly to the CFPS reaction, which was successful in the cell-free production of manganese peroxidase as well as horseradish peroxidase [23][24][27]. Since AaeUPO and other UPOs have at least one internal disulfide bond and other UPOs form functional dimers via disulfide bridges, it might also be beneficial to add protein disulfide isomerases like PDI or DsbC to CFPS reactions [23][27].
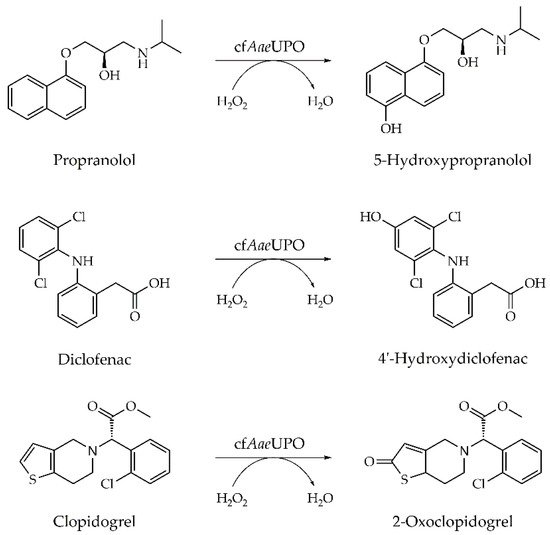

3.3. Substrate Screening with Cell-Free AaeUPO
The cell-free protein production approach should offer the possibility to establish a rapid screening platform for interesting biocatalysts. Thus, we tested the cell-free UPO produced with N. crassa lysate directly in a substrate screening without the need for time-consuming purification steps. To this end, the conversion (oxygenation, oxidation) of typical UPO substrates, which are also used in activity assays, as well as of three common pharmaceuticals was performed with simply prepared CFPS reaction solutions. The enzyme that is partly located in microsomes was efficiently released by ultrasonic treatment (Figure A3). After centrifugation, the supernatant was used for photometric UPO assays with ABTS, veratryl alcohol, 5-nitrobenzodioxole, and naphthalene, and for LC-MS analyses with the pharmaceuticals propranolol, diclofenac, and clopidogrel.
The β-adrenergic blocker propranolol, the non-steroidal anti-inflammatory drug diclofenac, and the antithrombotic prodrug clopidogrel are known to be metabolized by cytochrome P450 monooxygenases in the human body. In previous studies, it was shown that wtAaeUPO is able to perform the same reactions to give the (major) human drug metabolites 5-hydroxypropranolol, 4′-hydroxydiclofenac, and 2-oxoclopidogrel, respectively (Figure 5) [15][37][50].

Figure 5. Conversion of the pharmaceuticals propranolol, diclofenac, and clopidogrel by cell-free prepared UPO (cfAaeUPO).
The activities of cfAaeUPO were compared with those of its wild-type counterpart (taking into account that wtAaeUPO did not contain a polyhistidine tag). For the purpose of comparison, we used enzyme samples with a normalized volume activity of 100 U L−1 (based on veratryl alcohol oxidation) in all reactions. The results are listed in Table 2.
Table 2. Substrate conversion by homologous wtAaeUPO and cfAaeUPO. All reactions contained a UPO activity of 100 mU mL−1 (based on the oxidation of veratryl alcohol). The enzyme activities are given in mU mL−1 (a) or µU mL−1 (b).

All substrates were converted by both enzyme preparations, although quantitative differences in the activities were found in some cases. For NBD, naphthalene, and propranolol, the measured activities were similar; however, for ABTS, diclofenac, and clopidogrel, the activities of the cell-free variant were two to four times lower. ABTS is a substrate that is oxidized via one-electron abstraction by the peroxidative activity of UPO resulting in a cation radical product (‘classic’ peroxidase activity without oxygen transfer). The fungal lysate used for CFPS may have contained metabolites with antioxidant properties that could have interfered with the detection of the ABTS radical [51]. Thus, substrates like ABTS might only be suitable in screening approaches with unpurified UPO, if the yield of cell-free enzyme is relatively high. A possible explanation for the lower conversion of diclofenac and clopidogrel could be the presence of the C-terminal polyhistidine tag, which may have impaired activity, e.g., by shielding the heme-access channel. Such an effect was already reported for a mutant variant of AaeUPO (PaDa-I) [52]. In addition, negative effects of potentially different glycosylation patterns cannot be excluded.
In summary, it took less than two days from the DNA template carrying the AaeUPO gene to obtain the substrate screening results with expressed cfAaeUPO. This is much faster compared to conventional screening systems based on heterologous expression in host microbes [53]. Moreover, if transcription and translation were coupled in one CFPS reaction, the turnaround time of the entire process could be reduced even further.
4. Conclusions
In the present study, we evaluated a novel approach to produce unspecific peroxygenases (UPOs) by using a cell-free synthesis system based on fungal lysates. The UPO from Cyclocybe (Agrocybe) aegerita was successfully expressed with lysates derived from N. crassa or A. niger. Activities of this cfAaeUPO of up to 105 U L−1 (based on veratryl alcohol oxidation) could be achieved within 24 h, where the lysate from N. crassa showed a significantly higher translational performance compared to A. niger. The unpurified CFPS reaction solutions were subjected to a substrate screening, in which the performance of the cell-free enzyme (cfAaeUPO) was comparable to that of the homologously produced enzyme (wtAaeUPO). The procedure presented here highlights the general potential of CFPS for rapid screening of new UPOs or UPO variants, including the estimation of their catalytic performance. Nevertheless, for large-scale production of suitable candidates, hetero-logous production in fungal hosts will continue to be the method of choice, as cell-free synthesis is currently not competitive in terms of yield. Last, but not least, it must be pointed out that the full potential of lysates from filamentous fungi is far from being tapped, as their use in CFPS is still in its infancy.
References
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic synthesis for industrial applications. Angew. Chem. Int. Ed. 2021, 60, 88–119.
- Zhang, Z.-J.; Pan, J.; Ma, B.-D.; Xu, J.-H. Efficient biocatalytic synthesis of chiral chemicals. In Bioreactor Engineering Research and Industrial Applications I; Ye, Q., Bao, J., Zhong, J.-J., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 155, pp. 55–106.
- Sheldon, R.A.; Brady, D.; Bode, M.L. The Hitchhiker’s guide to biocatalysis: Recent advances in the use of enzymes in organic synthesis. Chem. Sci. 2020, 11, 2587–2605.
- Hofrichter, M.; Kellner, H.; Pecyna, M.J.; Ullrich, R. Fungal unspecific peroxygenases: Heme-thiolate proteins that combine peroxidase and cytochrome P450 properties. In Monooxygenase, Peroxidase and Peroxygenase Properties and Mechanisms of Cytochrome P450; Hrycay, E.G., Bandiera, S.M., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; Volume 851, pp. 341–368.
- Hofrichter, M.; Kellner, H.; Herzog, R.; Karich, A.; Kiebist, J.; Scheibner, K.; Ullrich, R. Peroxide-mediated oxygenation of organic compounds by fungal peroxygenases. Antioxidants 2022, 11, 163.
- Wang, Y.; Lan, D.; Durrani, R.; Hollmann, F. Peroxygenases en route to becoming dream catalysts. What are the opportunities and challenges? Curr. Opin. Chem. Biol. 2017, 37, 1–9.
- Sigmund, M.-C.; Poelarends, G.J. Current state and future perspectives of engineered and artificial peroxygenases for the oxyfunctionalization of organic molecules. Nat. Catal. 2020, 3, 690–702.
- Kiebist, J.; Hofrichter, M.; Zuhse, R.; Scheibner, K. Oxyfunctionalization of pharmaceuticals by fungal peroxygenases. In Pharmaceutical Biocatalysis, 1st ed.; Grunwald, P., Ed.; Jenny Stanford Publishing: Singapore, 2019; pp. 643–680.
- Hobisch, M.; Holtmann, D.; Gomez de Santos, P.; Alcalde, M.; Hollmann, F.; Kara, S. Recent developments in the use of peroxygenases—Exploring their high potential in selective oxyfunctionalisations. Biotechnol. Adv. 2021, 51, 107615.
- Kiebist, J.; Holla, W.; Heidrich, J.; Poraj-Kobielska, M.; Sandvoss, M.; Simonis, R.; Gröbe, G.; Atzrodt, J.; Hofrichter, M.; Scheibner, K. One-pot synthesis of human metabolites of SAR548304 by fungal peroxygenases. Bioorg. Med. Chem. 2015, 23, 4324–4332.
- Steinbrecht, S.; Kiebist, J.; König, R.; Thiessen, M.; Schmidtke, K.-U.; Kammerer, S.; Küpper, J.-H.; Scheibner, K. Synthesis of cyclophosphamide metabolites by a peroxygenase from Marasmius rotula for toxicological studies on human cancer cells. AMB Express 2020, 10, 128.
- Gomez de Santos, P.; Cañellas, M.; Tieves, F.; Younes, S.H.H.; Molina-Espeja, P.; Hofrichter, M.; Hollmann, F.; Guallar, V.; Alcalde, M. Selective synthesis of the human drug metabolite 5′-hydroxypropranolol by an evolved self-sufficient peroxygenase. ACS Catal. 2018, 8, 4789–4799.
- Kiebist, J.; Schmidtke, K.-U.; Zimmermann, J.; Kellner, H.; Jehmlich, N.; Ullrich, R.; Zänder, D.; Hofrichter, M.; Scheibner, K. A peroxygenase from Chaetomium globosum catalyzes the selective oxygenation of testosterone. Chembiochem 2017, 18, 563–569.
- Ullrich, R.; Poraj-Kobielska, M.; Scholze, S.; Halbout, C.; Sandvoss, M.; Pecyna, M.J.; Scheibner, K.; Hofrichter, M. Side chain removal from corticosteroids by unspecific peroxygenase. J. Inorg. Biochem. 2018, 183, 84–93.
- Kiebist, J.; Schmidtke, K.-U.; Schramm, M.; König, R.; Quint, S.; Kohlmann, J.; Zuhse, R.; Ullrich, R.; Hofrichter, M.; Scheibner, K. Biocatalytic syntheses of antiplatelet metabolites of the thienopyridines clopidogrel and prasugrel using fungal peroxygenases. J. Fungi 2021, 7, 752.
- Ullrich, R.; Karich, A.; Hofrichter, M. Fungal peroxygenases—A versatile tool for biocatalysis. In Encyclopedia of Mycology; Zaragoza, Ó., Casadevall, A., Eds.; Elsevier: Oxford, UK, 2021; pp. 260–280.
- Ullrich, R.; Nüske, J.; Scheibner, K.; Spantzel, J.; Hofrichter, M. Novel haloperoxidase from the agaric basidiomycete Agrocybe aegerita oxidizes aryl alcohols and aldehydes. Appl. Environ. Microbiol. 2004, 70, 4575–4581.
- Anh, D.H.; Ullrich, R.; Benndorf, D.; Svatoś, A.; Muck, A.; Hofrichter, M. The coprophilous mushroom Coprinus radians secretes a haloperoxidase that catalyzes aromatic peroxygenation. Appl. Environ. Microbiol. 2007, 73, 5477–5485.
- Gröbe, G.; Ullrich, R.; Pecyna, M.J.; Kapturska, D.; Friedrich, S.; Hofrichter, M.; Scheibner, K. High-yield production of aromatic peroxygenase by the agaric fungus Marasmius rotula. AMB Express 2011, 1, 31.
- Hofrichter, M.; Kellner, H.; Herzog, R.; Karich, A.; Liers, C.; Scheibner, K.; Kimani, V.W.; Ullrich, R. Fungal peroxygenases: A phylogenetically old superfamily of heme enzymes with promiscuity for oxygen transfer reactions. In Grand Challenges in Fungal Biotechnology; Nevalainen, H., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 369–403.
- Kinner, A.; Rosenthal, K.; Lütz, S. Identification and expression of new unspecific peroxygenases—Recent advances, challenges and opportunities. Front. Bioeng. Biotechnol. 2021, 9, 705630.
- Rotilio, L.; Swoboda, A.; Ebner, K.; Rinnofner, C.; Glieder, A.; Kroutil, W.; Mattevi, A. Structural and biochemical studies enlighten the unspecific peroxygenase from Hypoxylon sp. EC38 as an efficient oxidative biocatalyst. ACS Catal. 2021, 11, 11511–11525.
- Miyazaki-Imamura, C.; Oohira, K.; Kitagawa, R.; Nakano, H.; Yamane, T.; Takahashi, H. Improvement of H2O2 stability of manganese peroxidase by combinatorial mutagenesis and high-throughput screening using in vitro expression with protein disulfide isomerase. Protein Eng. Des. Sel. 2003, 16, 423–428.
- Ninomiya, R.; Zhu, B.; Kojima, T.; Iwasaki, Y.; Nakano, H. Role of disulfide bond isomerase DsbC, calcium ions, and hemin in cell-free protein synthesis of active manganese peroxidase isolated from Phanerochaete chrysosporium. J. Biosci. Bioeng. 2014, 117, 652–657.
- Kwon, Y.-C.; Oh, I.-S.; Lee, N.; Lee, K.-H.; Yoon, Y.J.; Lee, E.Y.; Kim, B.-G.; Kim, D.-M. Integrating cell-free biosyntheses of heme prosthetic group and apoenzyme for the synthesis of functional P450 monooxygenase. Biotechnol. Bioeng. 2013, 110, 1193–1200.
- Katayama, Y.; Shimokata, K.; Suematsu, M.; Ogura, T.; Tsukihara, T.; Yoshikawa, S.; Shimada, H. Cell-free synthesis of cytochrome c oxidase, a multicomponent membrane protein. J. Bioenerg. Biomembr. 2010, 42, 235–240.
- Zhu, B.; Mizoguchi, T.; Kojima, T.; Nakano, H. Ultra-high-throughput screening of an in vitro-synthesized horseradish peroxidase displayed on microbeads using cell sorter. PLoS ONE 2015, 10, e0127479.
- Rolf, J.; Rosenthal, K.; Lütz, S. Application of cell-free protein synthesis for faster biocatalyst development. Catalysts 2019, 9, 190.
- Gregorio, N.E.; Levine, M.Z.; Oza, J.P. A user’s guide to cell-free protein synthesis. Methods Protoc. 2019, 2, 24.
- Szczin-Skorupa, E.; Filipowicz, W.; Paszewski, A. The cell-free protein synthesis system from the ‘slime’ mutant of Neurospora crassa. Preparation and characterisation of importance of 7-methylguanosine for translation of viral and cellular mRNAs. Eur. J. Biochem. 1981, 121, 163–168.
- Yu, C.-H.; Dang, Y.; Zhou, Z.; Wu, C.; Zhao, F.; Sachs, M.S.; Liu, Y. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell 2015, 59, 744–754.
- Wang, Z.; Sachs, M.S. Arginine-specific regulation mediated by the Neurospora crassa arg-2 upstream open reading frame in a homologous, cell-free in vitro translation system. J. Biol. Chem. 1997, 272, 255–261.
- Wu, C.; Dasgupta, A.; Shen, L.; Bell-Pedersen, D.; Sachs, M.S. The cell free protein synthesis system from the model filamentous fungus Neurospora crassa. Methods 2018, 137, 11–19.
- Devchand, M.; Gwynne, D.; Buxton, F.P.; Davies, R.W. An efficient cell-free translation system from Aspergillus nidulans and in vitro translocation of prepro-a-factor across Aspergillus microsomes. Curr. Genet. 1988, 14, 561–566.
- Gan, R.; Jewett, M.C. A combined cell-free transcription-translation system from Saccharomyces cerevisiae for rapid and robust protein synthe. Biotechnol. J. 2014, 9, 641–651.
- Pecyna, M.J.; Ullrich, R.; Bittner, B.; Clemens, A.; Scheibner, K.; Schubert, R.; Hofrichter, M. Molecular characterization of aromatic peroxygenase from Agrocybe aegerita. Appl. Microbiol. Biotechnol. 2009, 84, 885–897.
- Kinne, M.; Poraj-Kobielska, M.; Aranda, E.; Ullrich, R.; Hammel, K.E.; Scheibner, K.; Hofrichter, M. Regioselective preparation of 5-hydroxypropranolol and 4′-hydroxydiclofenac with a fungal peroxygenase. Bioorg. Med. Chem. Lett. 2009, 19, 3085–3087.
- Thoring, L.; Dondapati, S.K.; Stech, M.; Wüstenhagen, D.A.; Kubick, S. High-yield production of “difficult-to-express” proteins in a continuous exchange cell-free system based on CHO cell lysates. Sci. Rep. 2017, 7, 11710.
- Lichty, J.J.; Malecki, J.L.; Agnew, H.D.; Michelson-Horowitz, D.J.; Tan, S. Comparison of affinity tags for protein purification. Protein Expr. Purif. 2005, 41, 98–105.
- Hodgman, C.E.; Jewett, M.C. Optimized extract preparation methods and reaction conditions for improved yeast cell-free protein synthesis: Improved Yeast CFPS. Biotechnol. Bioeng. 2013, 110, 2643–2654.
- Schoborg, J.A.; Hodgman, C.E.; Anderson, M.J.; Jewett, M.C. Substrate replenishment and byproduct removal improve yeast cell-free protein synthesis. Biotechnol. J. 2014, 9, 630–640.
- Molina-Espeja, P.; Garcia-Ruiz, E.; Gonzalez-Perez, D.; Ullrich, R.; Hofrichter, M.; Alcalde, M. Directed evolution of unspecific peroxygenase from Agrocybe aegerita. Appl. Environ. Microbiol. 2014, 80, 3496–3507.
- Molina-Espeja, P.; Ma, S.; Mate, D.M.; Ludwig, R.; Alcalde, M. Tandem-yeast expression system for engineering and producing unspecific peroxygenase. Enzyme Microb. Technol. 2015, 73–74, 29–33.
- Miguez, A.M.; McNerney, M.P.; Styczynski, M.P. Metabolic profiling of Escherichia coli -based cell-free expression systems for process optimization. Ind. Eng. Chem. Res. 2019, 58, 22472–22482.
- Borkowski, O.; Koch, M.; Zettor, A.; Pandi, A.; Batista, A.C.; Soudier, P.; Faulon, J.-L. Large scale active-learning-guided exploration for in vitro protein production optimization. Nat. Commun. 2020, 11, 1872.
- Hodgman, C.E.; Jewett, M.C. Characterizing IGR IRES-mediated translation initiation for use in yeast cell-free protein synthesis. New Biotechnol. 2014, 31, 499–505.
- Le Fourn, V.; Girod, P.-A.; Buceta, M.; Regamey, A.; Mermod, N. CHO cell engineering to prevent polypeptide aggregation and improve therapeutic protein secretion. Metab. Eng. 2014, 21, 91–102.
- Stech, M.; Quast, R.B.; Sachse, R.; Schulze, C.; Wüstenhagen, D.A.; Kubick, S. A continuous-exchange cell-free protein synthesis system based on extracts from cultured insect cells. PLoS ONE 2014, 9, e96635.
- Krainer, F.W.; Capone, S.; Jäger, M.; Vogl, T.; Gerstmann, M.; Glieder, A.; Herwig, C.; Spadiut, O. Optimizing cofactor availability for the production of recombinant heme peroxidase in Pichia pastoris. Microb. Cell Fact. 2015, 14, 4.
- Poraj-Kobielska, M.; Atzrodt, J.; Holla, W.; Sandvoss, M.; Grobe, G.; Scheibner, K.; Hofrichter, M. Preparation of labeled human drug metabolites and drug-drug interaction-probes with fungal peroxygenases. J. Label. Compd. Rad. 2013, 56, 513–519.
- Makky, E.A.; AlMatar, M.; Mahmood, M.H.; Ting, O.W.; Qi, W.Z. Evaluation of the antioxidant and antimicrobial activities of ethyl acetate extract of Saccharomyces cerevisiae. Food Technol. Biotechnol. 2021, 59, 127–136.
- Bormann, S.; Burek, B.O.; Ulber, R.; Holtmann, D. Immobilization of unspecific peroxygenase expressed in Pichia pastoris by metal affinity binding. Mol. Catal. 2020, 492, 110999.
- Pullmann, P.; Knorrscheidt, A.; Munch, J.; Palme, P.R.; Hoehenwarter, W.; Marillonnet, S.; Alcalde, M.; Westermann, B.; Weissenborn, M.J. A modular two yeast species secretion system for the production and preparative application of unspecific peroxygenases. Commun. Biol. 2021, 4, 562.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
2 times
(View History)
Update Date:
07 Feb 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




