Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Víctor Andrés Arrieta González | + 2503 word(s) | 2503 | 2021-12-21 08:43:46 | | | |
| 2 | Peter Tang | Meta information modification | 2503 | 2022-01-21 08:00:15 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Arrieta González, V. The Eclectic Nature of Glioma-Infiltrating Macrophages and Microglia. Encyclopedia. Available online: https://encyclopedia.pub/entry/18593 (accessed on 07 February 2026).
Arrieta González V. The Eclectic Nature of Glioma-Infiltrating Macrophages and Microglia. Encyclopedia. Available at: https://encyclopedia.pub/entry/18593. Accessed February 07, 2026.
Arrieta González, Víctor. "The Eclectic Nature of Glioma-Infiltrating Macrophages and Microglia" Encyclopedia, https://encyclopedia.pub/entry/18593 (accessed February 07, 2026).
Arrieta González, V. (2022, January 21). The Eclectic Nature of Glioma-Infiltrating Macrophages and Microglia. In Encyclopedia. https://encyclopedia.pub/entry/18593
Arrieta González, Víctor. "The Eclectic Nature of Glioma-Infiltrating Macrophages and Microglia." Encyclopedia. Web. 21 January, 2022.
Copy Citation
Glioblastomas (GBMs) are complex ecosystems composed of highly multifaceted tumor and myeloid cells capable of responding to different environmental pressures, including therapies. Recent studies have uncovered the diverse phenotypical identities of brain-populating myeloid cells. Differences in the immune proportions and phenotypes within tumors seem to be dictated by molecular features of glioma cells. Furthermore, increasing evidence underscores the significance of interactions between myeloid cells and glioma cells that allow them to evolve in a synergistic fashion to sustain tumor growth.
microglia
macrophages
gliomas
immunotherapy
1. Introduction
The idea of the central nervous system (CNS) being immunologically privileged was based on the misconception of an impenetrable and quiescent state. However, this long-standing concept has been challenged and refuted. This change in perspective regarding CNS immune reactivity derives in part from the findings of an unrecognized meningeal lymphatic vascular system in the CNS [1][2][3]. This has contributed to the understanding of the means that are used by the immune cells and macromolecules to be transported outside the brain. Additionally, contiguous communication between the skull bone marrow and the brain provides an avenue for active traffic of myeloid cells to respond under emerging disturbances such as brain tumors, to CNS homeostasis [4]. Brain tumors release signals into the cerebrospinal fluid which communicates with the skull bone marrow to instruct cranial hematopoiesis [5]. This recent evidence further underscores the dynamic and unique nature of the brain immune system.
Myeloid cells, constituting microglia and monocytes that give rise to macrophages and dendritic cells, conglomerate in gliomas to exert a number of simultaneous actions in defending or supporting tumor growth. The convergence of different myeloid cells from diverse hematopoietic organs in gliomas likely contributes to the tumor cell heterogeneity described in these brain tumors. Estimations consider that around 30–50% of the tumor content is represented by monocyte-derived macrophages (MDMs) and resident microglia [6][7]. The mechanisms used by MDMs and microglia to populate gliomas are influenced by their embryological origins, spatial localization, and functional nature. As part of the innate immune defense, myeloid cells engulf tumor components, present antigens, produce cytokines, and induce direct cytotoxic activities on damaged cells, pathogens, and tumor cells [8][9].
Single-cell technologies have enabled exquisite characterizations of the multidimensional phenotypic states of the myeloid cells in different brain geographic regions, in age, in health, and in the context of neurological diseases [10][11][12][13], including gliomas [14][15][16][17][18]. Glioma-infiltrating myeloid cells show a wide range of transcriptional phenotypes derived from the presence of both MDMs and microglia [14][15][16]. Despite this evidence, many studies consider macrophages and microglia as single myeloid entities. Instead, the recognition of microglia and MDMs as different entities as well as the heterogeneity of each of these immune cell populations in the context of brain tumors will clarify the mechanisms of response and resistance to immunotherapies attributed to these cells.
2. Dissecting Myeloid Cells in Gliomas: Multiple Actors Come into Play
Several differences between microglia and MDMs have been described comprehensively in gliomas and other neurological diseases. Particularly, microglia are immune cells derived from primitive yolk sac macrophages that arise during early embryogenesis [19][20]. This brain-specific type of cell was preserved through the evolution of species to maintain CNS homeostasis, shape the neuronal networks, and cope with pathogens and insults of different origins. Though some microglia display remarkable longevity [21], the pool of these immune cells is sustained by modest local expansion throughout adult life with different proliferative rates depending on the brain region [22]. During neuropathological conditions, specific microglial clones expand to contend with CNS damage and subsequently decrease in cell number upon returning to homeostatic states [22]. Although little is known about the proliferation dynamics of microglia under the influence of gliomas, microglia and MDM from gliomas possess the proliferating capacity that was assessed by Ki-67 expression [23][24]. This suggests that like other neurological diseases, microglial cells respond to oncogenic insults through proliferation and phenotypic changes.
Apart from the microglia, specialized subsets of macrophages exist in the CNS borders including the perivascular, leptomeningeal, choroid plexus, and dural niches [25]. CNS-associated macrophages are diverse and long-lived. Some of these cell populations are replenished via local self-renewal and others by bone-marrow macrophages [26][27]. The modulation and influence of these macrophages by gliomas is an area of increasing investigation. The other abundant macrophage population infiltrating gliomas emanate from peripheral monocytes which are generated from hematopoietic progenitors in the bone marrow [6], Figure 1. Importantly, the skull and the vertebral bone marrow are prominent sources of monocytes that travel through vascular channels to settle in the meninges and the brain parenchyma under homeostatic and neuroinflammatory conditions [4][28]. Considering this new evidence, it is reasonable to infer that monocyte progenitors derived from skull bone marrow contribute to the supply of MDMs in the tumor microenvironment (TME) of gliomas. However, the extent of this contribution has not yet been defined. The peripheral immune compartment of glioma patients is also a source of tumor-infiltrating myeloid cells. Particularly, relevant numbers of myeloid-derived suppressor cells (MDSCs) were found in matched peripheral blood and tumors of patients with GBM [29][30], the most aggressive type of gliomas classified as WHO grade 4 tumors [31]. MDSCs are myeloid progenitors at earlier stages of differentiation that develop and accumulate systemically and in the TME where they arrive to promote an immunosuppressive milieu in support of gliomagenesis [32][33].
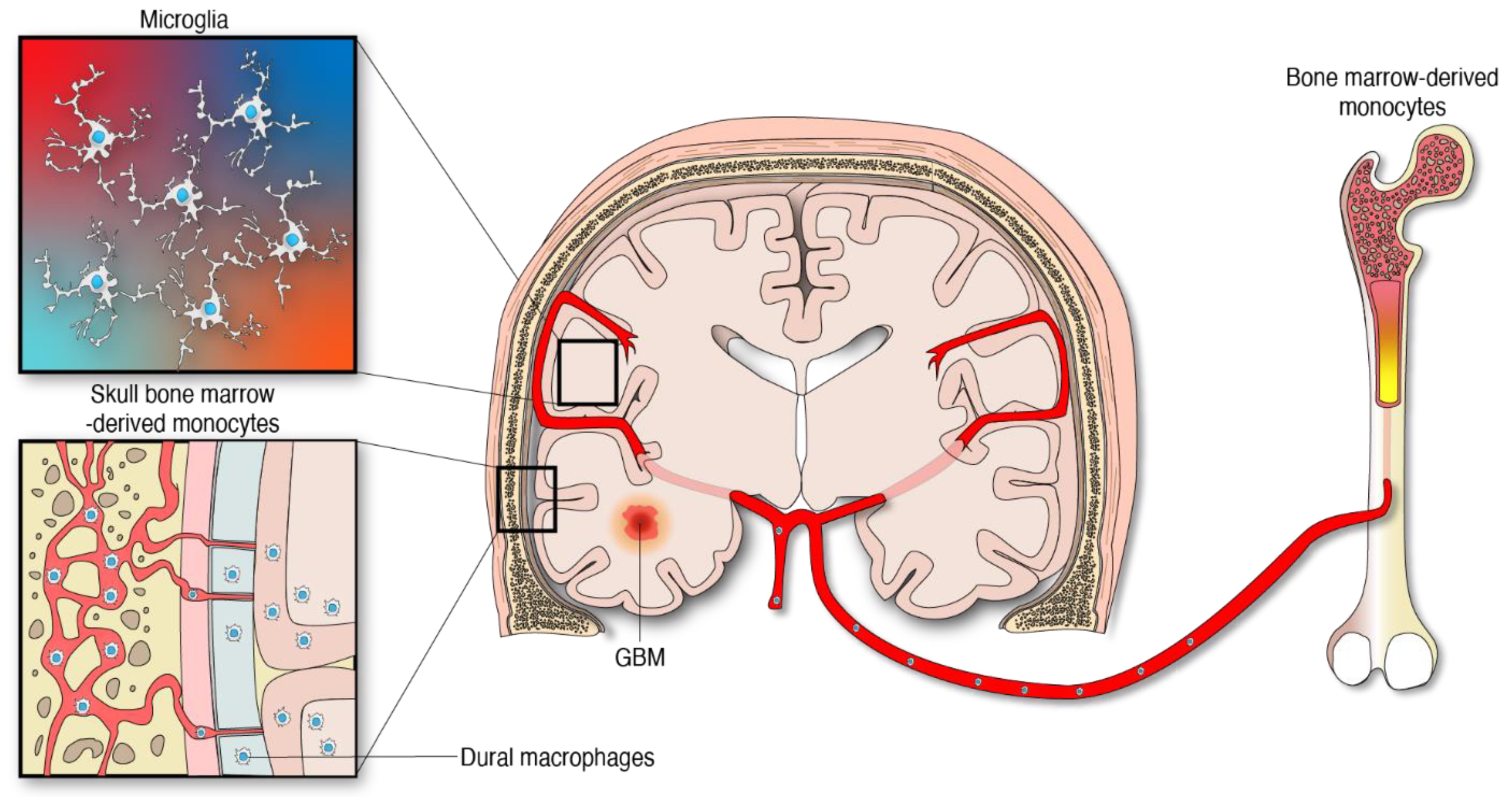
Figure 1. Myeloid cells from different anatomical and embryological origins contribute to the cellular heterogeneity in gliomas.
3. Differentiation Trajectories of Myeloid Cells under the Influence of Gliomas
The variability in immune activation and suppressive components among gliomas depends on the subtype and their localization in the brain [17][34]. In addition to these factors, newly diagnosed and recurrent GBMs differ in their immune composition and diversity due to the natural course of tumor progression and chemoradiotherapy [18]. Infiltration of peripheral monocyte/macrophages occurs in the early phases of tumor development and is maintained by constant immune cell recruitment throughout tumor progression as shown in murine glioma models [35][36]. This phenomenon is reflected in human gliomas where there are higher expression levels of MDMs genes compared to microglial genes in advanced stages of brain tumor disease irrespective of IDH mutational status [14][37]. From a different angle, microglia comprise the major proportion of myeloid cells in newly diagnosed GBM; whereas MDMs represent the most abundant myeloid cell population in recurrent GBMs [18]. The shift in the predominance of the myeloid cell population from microglia to MDMs suggests that these immune cells may compete for space in gliomas. Indeed, it was documented that the blockade of monocyte tumor infiltration results in a consequential increase in microglial cell numbers [18][38]. The spatial distribution of MDMs and microglia is represented in patches and interspersed between glioma cells throughout the entire tumor. These observations contrast with the notion of microglia being predominantly present at the tumor margin, a phenomenon seen in transplantable mouse glioma models [18]. Despite indicators of competition between microglia and MDMs in gliomas, many more immune cell populations are recruited and coexist in the complex ecosystem of gliomas.
The previous paradigm of categorizing glioma-infiltrating myeloid cells into M1 (anti-tumor, pro-inflammatory) and M2 (pro-tumor, immunosuppressive), whereas simple and convenient, does not represent the phenotypic diversity of these myeloid cells in the brain. Furthermore, the application of a nomenclature derived from in vitro functional characterization has led to a misunderstanding of the specific functions of glioma-infiltrating myeloid cells that is only now beginning to be clarified with single-cell sequencing. The multifaceted transcriptional phenotypes of myeloid cells uncovered in recent studies surpass the conventional M1/M2 classification as demonstrated by the following observations: a diverse expression of phenotypical states, mutually exclusive transcriptional programs when comparing glioma-infiltrating macrophages, lack of separation among these activated/alternative states, and co-expression of M1 and M2 markers such as IFN-γ and IL-4, among other markers by these immune cells [39][23]. Vast and compelling evidence arguing that the M1/M2 macrophage classification should be reconsidered was eloquently reviewed elsewhere [40]. Most importantly, this nomenclature appears misleading in terms of the semantics and the understanding of the cellular myeloid mosaic in GBM. Instead, the notion of multidimensional states of macrophages was suggested considering the spectrum of phenotypes displayed by macrophages under a wide variety of stimuli [41]. In the context of human gliomas, this phenomenon is illustrated by bioinformatic inferences of differentiation trajectories using single-cell technologies. As an example, the process of monocyte to macrophage transition was delineated by proteomic fate mapping in IDH wild-type and mutant gliomas [37]. This analysis showed three phenotype trajectories that monocytes follow upon invasion of glioma tissues. Single-cell RNA-seq analysis of GBM-infiltrating MDMs revealed a variety of macrophage clusters including: (1) a monocyte–macrophage transition; (2) phagocytic active/high lipid metabolism; (3) hypoxic/glycolysis active; (4) SEPP1low/ microglia-like; (5) SEPP1high/anti-inflammatory cluster and a (6) IFN-induced signature cluster [18]. The existence of these clusters suggests that upon invasion of glioma tissues, monocytes differentiate into macrophages and acquire specialized functions. In contrast with the idea of a continuum between two phenotypes, this further highlights the multiplicity of possibilities in which these leukocytes can transform in the context of brain malignancy, Figure 2. Furthermore, these analyses show multiple myeloid cell functions occurring simultaneously in the TME of gliomas.
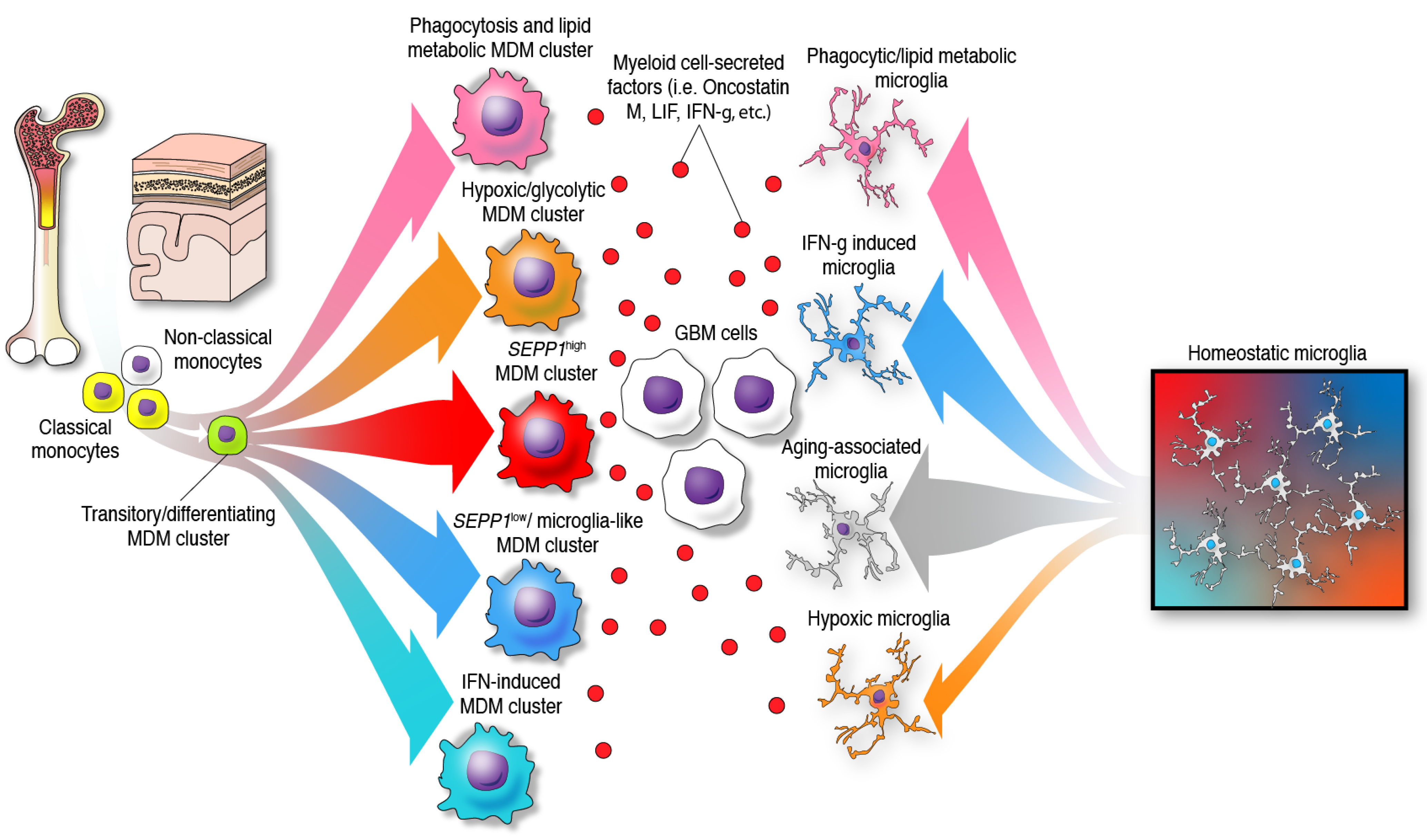
Figure 2. Identity and differentiation trajectories of MDMs and microglia under the influence of GBM cells in the TME. In the presence of gliomas, monocytes and microglia undergo differentiation towards different specific phenotypes.
4. Relationships and Interactions between the Tumor Genetics and Immune Landscape in Gliomas
The concept of genomic profile shaping immune responses in the TME and potential response to therapies is steadily gaining interest in neuro-oncology. Based on the differences in clinical behavior displayed by glioma patients and their distinctive tumor transcriptional profiles [42][43], IDH1/2 mutations were integrated into the molecular classification of these tumors [44]. In this regard, comprehensive characterizations of the TME of brain metastases, IDH wild-type and mutant gliomas were performed [23][37]. These studies have shown that IDH wild-type gliomas have a different myeloid cell composition and phenotype compared to those infiltrating IDH mutant gliomas. For instance, IDH wild-type gliomas were predominantly infiltrated by MDMs in contrast to IDH mutant gliomas which were characterized as harboring higher numbers of microglial cells. However, IDH wild-type gliomas had more microglial cells than brain metastases. Some of these differences can be attributed to the infiltrating versus focal nature of these malignancies. On the other hand, although not abundant, monocytes represented the main myeloid cell population derived from the blood in IDH mutant gliomas, indicating poor recruitment and deficient transition to tissue macrophages as compared to IDH wild-type gliomas that are abundantly infiltrated by differentiated MDMs [37]. There are other differences in cellular contents among IDH mutant gliomas including astrocytomas having high expression of microglia/macrophages gene signatures whereas oligodendrogliomas have enrichment of neuronal genes [14]. IDH mutant gliomas are less infiltrated by T cells and have lower expression of the PD-1 ligand, PD-L1, compared to their wild-type counterpart [45]. The T cell immune suppression in these gliomas is partially attributed to the presence of the oncometabolite (R)-2-hydroxyglutarate generated by the neomorphic activity of the IDH mutant enzyme [46]. Although not comparable with the relatively high number of lymphocytes in brain metastases, among IDH wild-type GBMs, a few have considerable levels of T cell infiltration which suggests that additional factors contribute to the differences in the composition of the lymphoid compartment among IDH wild-type tumors [23].
Although these previous studies have characterized the identity of immune infiltrates in IDH wild-type and mutant gliomas, the causal relationship between IDH genetic status and tumor-infiltrating immune cells remains a subject for future studies. Furthermore, recent studies characterizing the transcriptional profile of GBM have revealed subtypes associated with specific genetic abnormalities in cancer cells and the immune cellular content. They also provided important insights into the interactions between tumor and immune cells [47]. Compared to proneural and classical subtypes, GBMs classified as mesenchymal contained higher numbers of immune cells including lymphocytes, MDMs and microglia [34][48]. More recently, the depth of genomic deconvolution in GBM has allowed the identification of four cellular states that evolve through time and in accordance with distinct environmental cues. Mesenchymal-like (MES-like), astrocyte-like (AC-like), oligodendrocyte precursor cell-like (OPC-like) and neural progenitor cell-like (NPC-like) states represent these four modifiable oncogenic states described for GBM [49]. Shared biological processes employed during neurodevelopment underpin the expression programs of these cellular states except for the MES-like state which seems to be shaped in part by macrophages and T cell cytotoxic activity [39]. In the context of tumor progression-associated immune changes, innate immune cells contribute to the tumor mesenchymal differentiation predominantly in IDH wild-type GBM [50][51]. Upon brain infiltration, macrophages release several cytokines and ligands such as oncostatin M, which binds to the oncostatin M receptor and leukemia inhibitory factor receptor that signals through the signal transducer and activator of transcript 3 pathway to induce the MES-like transcriptional program in glioma cells [39]. This includes the potential of oncostatin M to induce the expression of MHC class I and II by glioma cells which results in increased susceptibility to T cell killing as shown by coculture assays [39]. The influence of macrophages on directing GBM cells towards a particular transcriptional program is also demonstrated by the effect of IFN-γ derived from these immune cells. Secreted IFN-γ induces the expression of immune-related gene signatures such as antigen processing and presentation, response to IFN-γ, response to IFN type I, among others by tumor cells [52]. Overall, these are examples that emphasize the importance of the interaction between tumor cells and the TME to shape the transcriptional and immune identity of glioma cells.
Of note, the mesenchymal signature can be induced without the presence of macrophages and induced by other internal and external cues [51]. On the other hand, considering that the recruitment of MDMs is a consequence of gliomagenesis, the genetic code of brain tumor cells ultimately determines the interaction with these myeloid cells from the beginning of cancer to evolve in a specified direction. For instance, genetic mutations or deletions of NF1 and PTEN, which are common alterations in mesenchymal GBM, appear to be responsible for the recruitment of MDMs and microglia [34]. Upon macrophage infiltration, the dialogue between myeloid cells and glioma cells through ligand-receptor interactions helps both cell types to thrive and synergize in the TME.
5. Glioma-Infiltrating Myeloid Cells under Therapy
Glioma-infiltrating myeloid cells are influenced by the standard of care consisting in chemoradiotherapy that GBM patients undergo. Radiotherapy induces substantial transcriptional changes in MDMs and microglia that are displayed when tumors recur. These gene expression changes result in a recurrence signature displayed by both MDMs and microglia [53]. Though a convergence in a transcriptional profile was found upregulated in myeloid cells, radiotherapy modified the myeloid cell abundance represented as an increase in MDMs and a decrease in microglia when tumors recurred in preclinical glioma models [53]. This is consistent with the change observed in the MDMs/microglia ratio between newly diagnosed and recurrent GBM patients [18][53].
GBM patients are often treated with dexamethasone to reduce tumor-associated cerebral edema and treatment-related adverse effects. However, steroid use is associated with worse survival and likely impairs the efficacy of anticancer therapies in GBM [54]. Furthermore, glucocorticoids are known to possess immunosuppressive properties. In fact, they were shown to profoundly affect MDMs and microglia, impairing monocyte tumor infiltration and inducing upregulation of a glucocorticoid gene signature in the myeloid cells isolated from newly diagnosed GBMs [18][55].
References
- Louveau, A.; Smirnov, I.; Keyes, T.J.; Eccles, J.D.; Rouhani, S.J.; Peske, J.D.; Derecki, N.C.; Castle, D.; Mandell, J.W.; Lee, K.S.; et al. Structural and functional features of central nervous system lymphatic vessels. Nature 2015, 523, 337–341.
- Aspelund, A.; Antila, S.; Proulx, S.T.; Karlsen, T.V.; Karaman, S.; Detmar, M.; Wiig, H.; Alitalo, K. A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J. Exp. Med. 2015, 212, 991–999.
- Louveau, A.; Herz, J.; Alme, M.N.; Salvador, A.F.; Dong, M.Q.; Viar, K.E.; Herod, S.G.; Knopp, J.; Setliff, J.C.; Lupi, A.L.; et al. CNS lymphatic drainage and neuroinflammation are regulated by meningeal lymphatic vasculature. Nat. Neurosci. 2018, 21, 1380–1391.
- Cugurra, A.; Mamuladze, T.; Rustenhoven, J.; Dykstra, T.; Beroshvili, G.; Greenberg, Z.J.; Baker, W.; Papadopoulos, Z.; Drieu, A.; Blackburn, S.; et al. Skull and vertebral bone marrow are myeloid cell reservoirs for the meninges and CNS parenchyma. Science 2021, 373, eabf7844.
- Pulous, F.E.; Cruz-Hernández, J.C.; Yang, C.; Kaya, Z.; Wojtkiewicz, G.; Capen, D.; Brown, D.; Wu, J.W.; Vinegoni, C.; Yamazoe, M.; et al. Cerebrospinal fluid outflow through skull channels instructs cranial hematopoiesis. bioRxiv 2021, 52, 3692–3695.
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016, 19, 20–27.
- Chen, Z.; Feng, X.; Herting, C.J.; Garcia, V.A.; Nie, K.; Pong, W.W.; Rasmussen, R.; Dwivedi, B.; Seby, S.; Wolf, S.A.; et al. Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma. Cancer Res. 2017, 77, 2266–2278.
- Michell-Robinson, M.A.; Touil, H.; Healy, L.M.; Owen, D.R.; Durafourt, B.A.; Bar-Or, A.; Antel, J.P.; Moore, C.S. Roles of microglia in brain development, tissue maintenance and repair. Brain 2015, 138, 1138–1159.
- Jaiswal, S.; Chao, M.P.; Majeti, R.; Weissman, I.L. Macrophages as mediators of tumor immunosurveillance. Trends Immunol. 2010, 31, 212–219.
- Masuda, T.; Sankowski, R.; Staszewski, O.; Böttcher, C.; Amann, L.; Sagar; Scheiwe, C.; Nessler, S.; Kunz, P.; van Loo, G.; et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 2019, 566, 388–392.
- Mathys, H.; Davila-Velderrain, J.; Peng, Z.; Gao, F.; Mohammadi, S.; Young, J.Z.; Menon, M.; He, L.; Abdurrob, F.; Jiang, X.; et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 2019, 570, 332–337.
- Farhadian, S.F.; Mehta, S.S.; Zografou, C.; Robertson, K.; Price, R.W.; Pappalardo, J.; Chiarella, J.; Hafler, D.A.; Spudich, S.S. Single-cell RNA sequencing reveals microglia-like cells in cerebrospinal fluid during virologically suppressed HIV. JCI Insight 2018, 3, e121718.
- Masuda, T.; Sankowski, R.; Staszewski, O.; Prinz, M. Microglia Heterogeneity in the Single-Cell Era. Cell Rep. 2020, 30, 1271–1281.
- Venteicher, A.S.; Tirosh, I.; Hebert, C.; Yizhak, K.; Neftel, C.; Filbin, M.G.; Hovestadt, V.; Escalante, L.E.; Shaw, M.L.; Rodman, C.; et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 2017, 355.
- Yuan, J.; Levitin, H.M.; Frattini, V.; Bush, E.C.; Boyett, D.M.; Samanamud, J.; Ceccarelli, M.; Dovas, A.; Zanazzi, G.; Canoll, P.; et al. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med. 2018, 10, 57.
- Müller, S.; Kohanbash, G.; Liu, S.J.; Alvarado, B.; Carrera, D.; Bhaduri, A.; Watchmaker, P.B.; Yagnik, G.; Di Lullo, E.; Malatesta, M.; et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 2017, 18, 234.
- Sankowski, R.; Böttcher, C.; Masuda, T.; Geirsdottir, L.; Sagar; Sindram, E.; Seredenina, T.; Muhs, A.; Scheiwe, C.; Shah, M.J.; et al. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat. Neurosci. 2019, 22, 2098–2110.
- Pombo Antunes, A.R.; Scheyltjens, I.; Lodi, F.; Messiaen, J.; Antoranz, A.; Duerinck, J.; Kancheva, D.; Martens, L.; De Vlaminck, K.; Van Hove, H.; et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci. 2021, 24, 595–610.
- Ginhoux, F.; Greter, M.; Leboeuf, M.; Nandi, S.; See, P.; Gokhan, S.; Mehler, M.F.; Conway, S.J.; Ng, L.G.; Stanley, E.R.; et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010, 330, 841–845.
- Kierdorf, K.; Erny, D.; Goldmann, T.; Sander, V.; Schulz, C.; Perdiguero, E.G.; Wieghofer, P.; Heinrich, A.; Riemke, P.; Hölscher, C.; et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 2013, 16, 273–280.
- Füger, P.; Hefendehl, J.K.; Veeraraghavalu, K.; Wendeln, A.C.; Schlosser, C.; Obermüller, U.; Wegenast-Braun, B.M.; Neher, J.J.; Martus, P.; Kohsaka, S.; et al. Microglia turnover with aging and in an Alzheimer’s model via long-term in vivo single-cell imaging. Nat. Neurosci. 2017, 20, 1371–1376.
- Tay, T.L.; Mai, D.; Dautzenberg, J.; Fernández-Klett, F.; Lin, G.; Sagar; Datta, M.; Drougard, A.; Stempfl, T.; Ardura-Fabregat, A.; et al. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci. 2017, 20, 793–803.
- Klemm, F.; Maas, R.R.; Bowman, R.L.; Kornete, M.; Soukup, K.; Nassiri, S.; Brouland, J.P.; Iacobuzio-Donahue, C.A.; Brennan, C.; Tabar, V.; et al. Interrogation of the Microenvironmental Landscape in Brain Tumors Reveals Disease-Specific Alterations of Immune Cells. Cell 2020, 181, 1643–1660.e1617.
- Chen, A.X.; Gartrell, R.D.; Zhao, J.; Upadhyayula, P.S.; Zhao, W.; Yuan, J.; Minns, H.E.; Dovas, A.; Bruce, J.N.; Lasorella, A.; et al. Single-cell characterization of macrophages in glioblastoma reveals MARCO as a mesenchymal pro-tumor marker. Genome Med. 2021, 13, 88.
- Kierdorf, K.; Masuda, T.; Jordão, M.J.C.; Prinz, M. Macrophages at CNS interfaces: Ontogeny and function in health and disease. Nat. Rev. Neurosci. 2019, 20, 547–562.
- Van Hove, H.; Martens, L.; Scheyltjens, I.; De Vlaminck, K.; Pombo Antunes, A.R.; De Prijck, S.; Vandamme, N.; De Schepper, S.; Van Isterdael, G.; Scott, C.L.; et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 2019, 22, 1021–1035.
- Mrdjen, D.; Pavlovic, A.; Hartmann, F.J.; Schreiner, B.; Utz, S.G.; Leung, B.P.; Lelios, I.; Heppner, F.L.; Kipnis, J.; Merkler, D.; et al. High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity 2018, 48, 380–395.e386.
- Herisson, F.; Frodermann, V.; Courties, G.; Rohde, D.; Sun, Y.; Vandoorne, K.; Wojtkiewicz, G.R.; Masson, G.S.; Vinegoni, C.; Kim, J.; et al. Direct vascular channels connect skull bone marrow and the brain surface enabling myeloid cell migration. Nat. Neurosci. 2018, 21, 1209–1217.
- Alban, T.J.; Alvarado, A.G.; Sorensen, M.D.; Bayik, D.; Volovetz, J.; Serbinowski, E.; Mulkearns-Hubert, E.E.; Sinyuk, M.; Hale, J.S.; Onzi, G.R.; et al. Global immune fingerprinting in glioblastoma patient peripheral blood reveals immune-suppression signatures associated with prognosis. JCI Insight 2018, 3, e122264.
- Gabrusiewicz, K.; Rodriguez, B.; Wei, J.; Hashimoto, Y.; Healy, L.M.; Maiti, S.N.; Thomas, G.; Zhou, S.; Wang, Q.; Elakkad, A.; et al. Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight 2016, 1.
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251.
- Kohanbash, G.; Okada, H. Myeloid-derived suppressor cells (MDSCs) in gliomas and glioma-development. Immunol. Investig. 2012, 41, 658–679.
- Fujita, M.; Kohanbash, G.; Fellows-Mayle, W.; Hamilton, R.L.; Komohara, Y.; Decker, S.A.; Ohlfest, J.R.; Okada, H. COX-2 blockade suppresses gliomagenesis by inhibiting myeloid-derived suppressor cells. Cancer Res. 2011, 71, 2664–2674.
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; deCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56.e46.
- Kennedy, B.C.; Maier, L.M.; D’Amico, R.; Mandigo, C.E.; Fontana, E.J.; Waziri, A.; Assanah, M.C.; Canoll, P.; Anderson, R.C.; Anderson, D.E.; et al. Dynamics of central and peripheral immunomodulation in a murine glioma model. BMC Immunol. 2009, 10, 11.
- Müller, A.; Brandenburg, S.; Turkowski, K.; Müller, S.; Vajkoczy, P. Resident microglia, and not peripheral macrophages, are the main source of brain tumor mononuclear cells. Int. J. Cancer 2015, 137, 278–288.
- Friebel, E.; Kapolou, K.; Unger, S.; Núñez, N.G.; Utz, S.; Rushing, E.J.; Regli, L.; Weller, M.; Greter, M.; Tugues, S.; et al. Single-Cell Mapping of Human Brain Cancer Reveals Tumor-Specific Instruction of Tissue-Invading Leukocytes. Cell 2020, 181, 1626–1642.e1620.
- Takenaka, M.C.; Gabriely, G.; Rothhammer, V.; Mascanfroni, I.D.; Wheeler, M.A.; Chao, C.C.; Gutiérrez-Vázquez, C.; Kenison, J.; Tjon, E.C.; Barroso, A.; et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat. Neurosci. 2019, 22, 729–740.
- Hara, T.; Chanoch-Myers, R.; Mathewson, N.D.; Myskiw, C.; Atta, L.; Bussema, L.; Eichhorn, S.W.; Greenwald, A.C.; Kinker, G.S.; Rodman, C.; et al. Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell 2021, 39, 779–792.e711.
- Ransohoff, R.M. A polarizing question: Do M1 and M2 microglia exist? Nat. Neurosci. 2016, 19, 987–991.
- Xue, J.; Schmidt, S.V.; Sander, J.; Draffehn, A.; Krebs, W.; Quester, I.; De Nardo, D.; Gohel, T.D.; Emde, M.; Schmidleithner, L.; et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014, 40, 274–288.
- Unruh, D.; Zewde, M.; Buss, A.; Drumm, M.R.; Tran, A.N.; Scholtens, D.M.; Horbinski, C. Methylation and transcription patterns are distinct in IDH mutant gliomas compared to other IDH mutant cancers. Sci. Rep. 2019, 9, 8946.
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773.
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820.
- Berghoff, A.S.; Kiesel, B.; Widhalm, G.; Wilhelm, D.; Rajky, O.; Kurscheid, S.; Kresl, P.; Wöhrer, A.; Marosi, C.; Hegi, M.E.; et al. Correlation of immune phenotype with IDH mutation in diffuse glioma. Neuro-Oncology 2017.
- Bunse, L.; Pusch, S.; Bunse, T.; Sahm, F.; Sanghvi, K.; Friedrich, M.; Alansary, D.; Sonner, J.K.; Green, E.; Deumelandt, K.; et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat. Med. 2018, 24, 1192–1203.
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110.
- Kaffes, I.; Szulzewsky, F.; Chen, Z.; Herting, C.J.; Gabanic, B.; Velázquez Vega, J.E.; Shelton, J.; Switchenko, J.M.; Ross, J.L.; McSwain, L.F.; et al. Human Mesenchymal glioblastomas are characterized by an increased immune cell presence compared to Proneural and Classical tumors. Oncoimmunology 2019, 8, e1655360.
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, 835–849.e821.
- Sa, J.K.; Chang, N.; Lee, H.W.; Cho, H.J.; Ceccarelli, M.; Cerulo, L.; Yin, J.; Kim, S.S.; Caruso, F.P.; Lee, M.; et al. Transcriptional regulatory networks of tumor-associated macrophages that drive malignancy in mesenchymal glioblastoma. Genome Biol. 2020, 21, 216.
- Schmitt, M.J.; Company, C.; Dramaretska, Y.; Barozzi, I.; Göhrig, A.; Kertalli, S.; Großmann, M.; Naumann, H.; Sanchez-Bailon, M.P.; Hulsman, D.; et al. Phenotypic Mapping of Pathologic Cross-Talk between Glioblastoma and Innate Immune Cells by Synthetic Genetic Tracing. Cancer Discov. 2021, 11, 754–777.
- Gangoso, E.; Southgate, B.; Bradley, L.; Rus, S.; Galvez-Cancino, F.; McGivern, N.; Güç, E.; Kapourani, C.A.; Byron, A.; Ferguson, K.M.; et al. Glioblastomas acquire myeloid-affiliated transcriptional programs via epigenetic immunoediting to elicit immune evasion. Cell 2021, 184, 2454–2470.e2426.
- Akkari, L.; Bowman, R.L.; Tessier, J.; Klemm, F.; Handgraaf, S.M.; de Groot, M.; Quail, D.F.; Tillard, L.; Gadiot, J.; Huse, J.T.; et al. Dynamic changes in glioma macrophage populations after radiotherapy reveal CSF-1R inhibition as a strategy to overcome resistance. Sci. Transl. Med. 2020, 12.
- Pitter, K.L.; Tamagno, I.; Alikhanyan, K.; Hosni-Ahmed, A.; Pattwell, S.S.; Donnola, S.; Dai, C.; Ozawa, T.; Chang, M.; Chan, T.A.; et al. Corticosteroids compromise survival in glioblastoma. Brain 2016, 139, 1458–1471.
- Herting, C.J.; Chen, Z.; Maximov, V.; Duffy, A.; Szulzewsky, F.; Shayakhmetov, D.M.; Hambardzumyan, D. Tumour-associated macrophage-derived interleukin-1 mediates glioblastoma-associated cerebral oedema. Brain 2019, 142, 3834–3851.
More
Information
Subjects:
Immunology; Oncology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
863
Revisions:
2 times
(View History)
Update Date:
21 Jan 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




