
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Wen Xie | + 2342 word(s) | 2342 | 2022-01-19 09:50:20 | | | |
| 2 | Beatrix Zheng | + 468 word(s) | 2810 | 2022-01-20 02:10:31 | | |
Video Upload Options
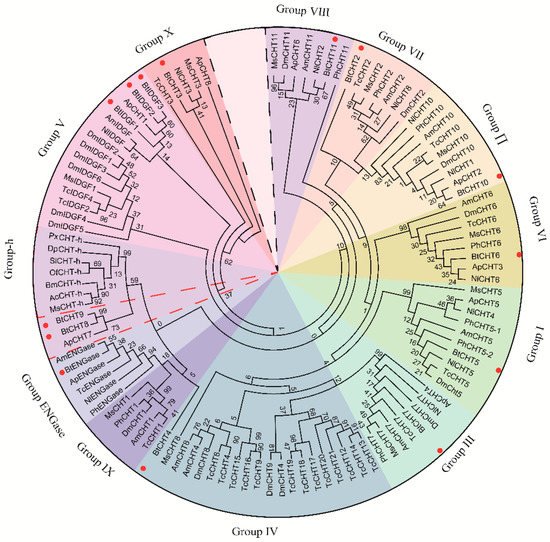
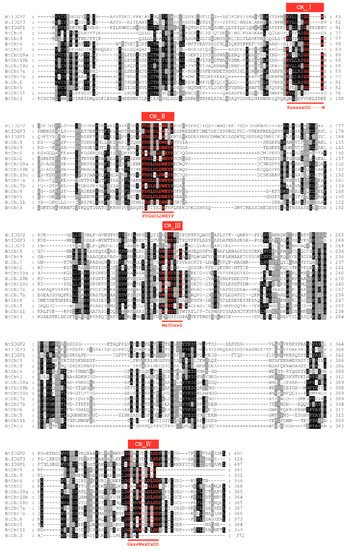
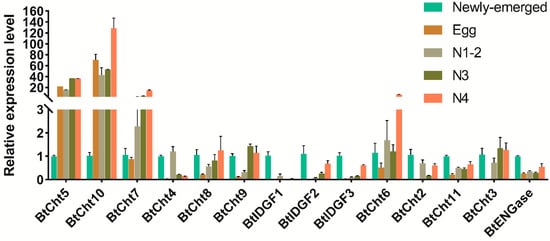
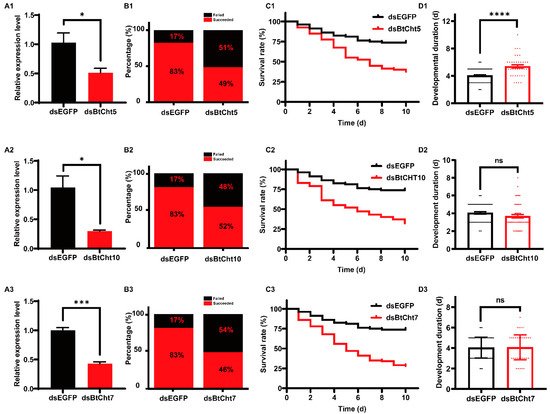
Chitinases are of great importance in chitin degradation and remodeling in insects. However, the genome-wide distribution of chitinase-like gene family in Bemsia tabaci, a destructive pest worldwide, is still elusive. With the help of bioinformatics, we annotated 14 genes that encode putative chitinase-like proteins, including ten chitinases (Cht), three imaginal disk growth factors (IDGF), and one endo-β-N-acetylglucosaminidase (ENGase) in the genome of the whitefly, B. tabaci. These genes were phylogenetically grouped into eight clades, among which 13 genes were classified in the glycoside hydrolase family 18 groups and one in the ENGase group. Afterwards, developmental expression analysis suggested that BtCht10, BtCht5, and BtCht7 were highly expressed in nymphal stages and exhibit similar expression patterns, implying their underlying role in nymph ecdysis. Notably, nymphs exhibited a lower rate of survival when challenged by dsRNA targeting these three genes via a nanomaterial-promoted RNAi method. In addition, silencing of BtCht10 significantly resulted in a longer duration of development compared to control nymphs. These results indicate a key role of BtCht10, BtCht5, and BtCht7 in B. tabaci nymph molting. This research depicts the differences of chitinase-like family genes in structure and function and identified potential targets for RNAi-based whitefly management.
1. Introduction
2. Current Insights
3. Conclusions
References
- Muthukrishnan, S.; Merzendorfer, H.; Arakane, Y.; Kramer, K.J. Chitin metabolism in insects. Insect Biochem. Mol. Biol. 2012, 193–235.
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206, 4393–4412.
- Zhu, Q.; Arakane, Y.; Banerjee, D.; Beeman, R.W.; Kramer, K.J.; Muthukrishnan, S. Domain organization and phylogenetic analysis of the chitinase-like family of proteins in three species of insects. Insect Biochem. Mol. Biol. 2008, 38, 452–466.
- Nakabachi, A.; Shigenobu, S.; Miyagishima, S. Chitinase-like proteins encoded in the genome of the pea aphid, Acyrthosiphon pisum. Insect Mol. Biol. 2010, 19, 175–185.
- Arakane, Y.; Muthukrishnan, S. Insect chitinase and chitinase-like proteins. Cell Mol. Life Sci. 2010, 67, 201–216.
- Funkhouser, J.D.; Aronson, N.N. Chitinase family GH18: Evolutionary insights from the genomic history of a diverse protein family. BMC Evol. Biol. 2007, 7, 96.
- Shuhui, L.; Mok, Y.K.; Wong, W.F. Role of mammalian chitinases in asthma. Int. Arch. Allergy Immunol. 2009, 149, 369–377.
- Kramer, K.J.; Muthukrishnan, S. Insect chitinases: Molecular biology and potential use as biopesticides. Insect Biochem. Mol. Biol. 1997, 27, 887–900.
- Fukamizo, T. Chitinolytic enzymes catalysis, substrate binding, and their application. Curr. Protein Pep. Sci. 2000, 1, 105–124.
- Zhu, Q.S.; Deng, Y.P.; Vanka, P.; Brown, S.J.; Muthukrishnan, S.; Kramer, K.J. Computational identification of novel chitinase-like proteins in the Drosophila melanogaster genome. Bioinformatics 2004, 20, 161–169.
- Tetreau, G.; Cao, X.; Chen, Y.R.; Muthukrishnan, S.; Jiang, H.; Blissard, G.W.; Kanost, M.R.; Wang, P. Overview of chitin metabolism enzymes in Manduca sexta: Identification, domain organization, phylogenetic analysis and gene expression. Insect Biochem. Mol. Biol. 2015, 62, 114–126.
- Zhu, Q.; Arakane, Y.; Beeman, R.W.; Kramer, K.J.; Muthukrishnan, S. Functional specialization among insect chitinase family genes revealed by RNA interference. Proc. Natl. Acad. Sci. USA 2008, 105, 6650–6655.
- Xi, Y.; Pan, P.L.; Ye, Y.X.; Yu, B.; Xu, H.J.; Zhang, C.X. Chitinase-like gene family in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 2015, 24, 29–40.
- Khajuria, C.; Buschman, L.L.; Chen, M.S.; Muthukrishnan, S.; Zhu, K.Y. A gut-specific chitinase gene essential for regulation of chitin content of peritrophic matrix and growth of Ostrinia nubilalis larvae. Insect Biochem. Mol. Biol. 2010, 40, 621–629.
- Cao, B.; Bao, W.; Wuriyanghan, H. Silencing of target chitinase genes via oral delivery of dsRNA caused lethal phenotypic effects in Mythimna separata (Lepidoptera: Noctuidae). Appl. Biochem. Biotechnol. 2017, 181, 860–866.
- Zhu, B.; Shan, J.Q.; Li, R.; Liang, P.; Gao, X.W. Identification and RNAi-based function analysis of chitinase family genes in diamondback moth, Plutella xylostella. Pest Manag. Sci. 2018, 75, 1951–1961.
- Zheng, Y.; Hu, Y.; Yan, S.; Zhou, H.; Song, D.; Yin, M.; Shen, J. A polymer/detergent formulation improves dsRNA penetration through the body wall and RNAi-induced mortality in the soybean aphid Aphis Glycines. Pest Manag. Sci. 2019, 75, 1993–1999.
- Shen, D.X.; Zhou, F.; Xu, Z.J.; He, B.C.; Li, M.; Shen, J.; Yin, M.Z.; An, C.J. Systemically interfering with immune response by a fluorescent cationic dendrimer delivered gene suppression. J. Mater. Chem. B 2014, 2, 4653–4659.
- Xu, Z.J.; He, B.C.; Wei, W.; Liu, K.L.; Yin, M.Z.; Yang, W.T.; Shen, J. Highly water-soluble perylenediimide-cored poly (amido amine) vector for efficient gene transfection. J. Mater. Chem. B 2014, 2, 4653–4659.
- Yan, S.; Ren, B.Y.; Shen, J. Nanoparticle-mediated double-stranded RNA delivery system: A promising approach for sustainable pest management. Insect Sci. 2021, 28, 21–34.
- De Barro, P.J.; Liu, S.S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A statement of species status. Annu. Rev. Entomol. 2011, 56, 1–19.
- Walling, L.L. Avoiding effective defenses: Strategies employed by phloem-feeding insects. Plant Physiol. 2008, 146, 859–866.
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu. Rev. Entomol. 2015, 2, 67–93.
- Dinsdale, A.; Cook, L.; Riginos, C.; Buckley, Y.M.; De Barro, P. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann. Entomol. Soc. Am. 2010, 103, 196–208.
- Boykin, L.M.; De Barro, P.J. A practical guide to identifying members of the Bemisia tabaci species complex: And other morphologically identical species. Front. Ecol. Evol. 2014, 2, 45.
- Ren, J.; Peng, Z.K.; Yang, Z.Z.; Tian, L.X.; Liu, S.N.; Wang, S.L.; Wu, Q.J.; Xie, W.; Zhang, Y.J. Genome-wide identification and analysis of sulfatase and sulfatase modifying factor genes in Bemisia tabaci (Hemiptera: Aleyrodidae). Insect Sci. 2020, 1–2.
- Su, Q.; Preisser, E.L.; Zhou, X.M.; Xie, W.; Liu, B.M.; Wang, S.L.; Wu, Q.J.; Zhang, Y.J. Manipulation of host quality and defense by a plant virus improves performance of whitefly vectors. J. Econ. Entomol. 2015, 108, 11–19.
- Basit, M. Status of insecticide resistance in Bemisia tabaci: Resistance, cross-resistance, stability of resistance, genetics and fitness costs. Phytoparasitica 2019, 47, 207–225.
- Horowitz, A.R.; Ghanim, M.; Roditakis, E.; Nauen, R.; Ishaaya, I. Insecticide resistance and its management in Bemisia tabaci species. J. Pest Sci. 2020, 93, 893–910.
- Li, S.J.; Ahmed, M.; Lv, N.; Shi, P.Q.; Wang, X.M.; Huang, J.L.; Qiu, B.L. Plant mediated horizontal transmission of Wolbachia between whiteflies. ISME J. 2017, 11, 1019–1028.
- Wang, S.L.; Zhang, Y.J.; Yang, X.; Xie, W.; Wu, Q.J. Resistance monitoring for eight insecticides on the sweetpotato whitefly (Hemiptera: Aleyrodidae) in China. J. Econ. Entomol. 2017, 110, 660–666.
- Nauen, R.; Denholm, I. Resistance of insect pests to neonicotinoid insecticides: Current status and future prospects. Arch. Ins. Biochem. Physiol. 2005, 58, 200–215.
- Roditakis, E.; Roditakis, N.E.; Tsagkarakou, A. Insecticide resistance in Bemisia tabaci (Homoptera: Aleyrodidae) populations from Crete. Pest Manag. Sci. 2005, 6, 577–582.
- Erdogan, C.; Moores, G.D.; Gurkan, M.O.; Gorman, K.J.; Denholm, I. Insecticide resistance and biotype status of populations of the tobacco whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) from Turkey. Crop Prot. 2008, 27, 600–605.
- Ahmad, M.; Arif, M.I.; Naveed, M. Dynamics of resistance to organophosphate and carbamate insecticides in the cotton whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) from Pakistan. J. Pest Sci. 2010, 83, 409–420.
- Luo, C.; Jones, C.M.; Devine, G.; Zhang, F.; Denholm, I.; Gorman, K. Insecticide resistance in Bemisia tabaci biotype Q (Hemiptera: Aleyrodidae) from China. Crop Prot. 2010, 29, 429–434.
- Wang, Z.; Yan, H.; Yang, Y.; Wu, Y. Biotype and insecticide resistance status of the whitefly Bemisia tabaci from China. Pest Manag. Sci. 2010, 66, 1360–1366.
- Vassiliou, V.; Emmanouilidou, M.; Perrakis, A.; Morou, E.; Vontas, J.; Tsagkarakou, A.; Roditakis, E. Insecticide resistance in Bemisia tabaci from Cyprus. Insect Sci. 2011, 18, 30–39.
- Kontsedalov, S.; Abu-Moch, F.; Lebedev, G.; Czosnek, H.; Horowitz, A.R.; Ghanim, M. Bemisia tabaci biotype dynamics and resistance to insecticides in Israel during the years 2008–2010. J. Integr. Agric. 2012, 11, 312–320.
- Peng, Z.K.; Zheng, H.X.; Xie, W.; Wang, S.L.; Wu, Q.J.; Zhang, Y.J. Field resistance monitoring of the immature stages of the whitefly Bemisia tabaci to spirotetramat in China. Crop Prot. 2017, 98, 243–247.
- Wang, R.; Che, W.; Wang, J.; Luo, C. Monitoring insecticide resistance and diagnostics of resistance mechanisms in Bemisia tabaci Mediterranean (Q biotype) in China. Pestic. Biochem. Phys. 2020, 163, 117–122.
- Chen, W.; Hasegawa, D.K.; Kaur, N.; Kliot, A.; Pinheiro, P.V.; Luan, J.B.; Marcus, C.; Stensmyr, M.C.; Zheng, Y.; Liu, W.L.; et al. The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 2016, 14, 1–15.
- Xie, W.; Chen, C.H.; Yang, Z.Z.; Guo, L.T.; Yang, X.X.; Wang, D.; Chen, M.; Huang, J.Q.; Wen, Y.N.; Zeng, Y.; et al. Genome sequencing of the sweetpotato whitefly Bemisia tabaci MED/Q. GigaScience 2017, 6, 1–7.
- Stansly, P.A.; Naranjo, S.E. (Eds.) Bemisia: Bionomics and Management of a Global Pest; Springer: New York, NY, USA, 2010; pp. 109–141.
- Bryant, P.J. Growth factors controlling imaginal disc growth in Drosophila. In The Cell Cycle and Development; Novartis Foundation Symposium; Wiley: New York, NY, USA, 2001; pp. 194–203.
- Varela, P.F.; Llera, A.S.; Mariuzza, R.A.; Tormo, J. Crystal structure of imaginal disc growth factor-2 a member of a new family of growth-promoting glycoproteins from Drosophila melanogaster. J. Biol. Chem. 2002, 277, 13229–13236.
- Broz, V.; Kucerova, L.; Rouhova, L.; Fleischmannova, J.; Strnad, H.; Bryant, P.J.; Zurovec, M. Drosophila imaginal disc growth factor 2 is a trophic factor involved in energy balance, detoxification, and innate immunity. Sci. Rep. 2017, 7, 43273.
- Gu, X.Y.; Li, Z.H.; Su, Y.; Zhao, Y.; Liu, L.J. Imaginal disc growth factor 4 regulates development and temperature adaptation in Bactrocera dorsalis. Sci. Rep. 2019, 9, 1–11.
- Matsuda, K.; Ihara, M.; Sattelle, D.B. Neonicotinoid insecticides: Molecular targets, resistance, and toxicity. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 241–255.
- Yang, X.; Deng, S.; Wei, X.G.; Yang, J.; Zhao, Q.N.; Yin, C.; Du, T.H.; Guo, Z.J.; Xia, J.X.; Yang, Z.Z.; et al. MAPK-directed activation of the whitefly transcription factor CREB leads to P450-mediated imidacloprid resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 10246–10253.
- Spindler, K.D.; Spindler-Barth, M.; Londershausen, M. Chitin metabolism: A target for drugs against parasites. Parasitol. Res. 1990, 76, 283–288.




