Non-Hodgkin lymphoma (NHL) is a heterogeneous malignancy with variable patient outcomes. There is still a lack of understanding about the different players involved in lymphomagenesis, and the identification of new diagnostic and prognostic biomarkers is urgent. MicroRNAs and long non-coding RNAs emerged as master regulators of B-cell development, and their deregulation has been associated with the initiation and progression of lymphomagenesis. They can function by acting alone or, as recently proposed, by creating competing endogenous RNA (ceRNA) networks. The study of miRNAs’ and lncRNAs’ deregulation in NHL, either alone or as ceRNAs networks, offers new insights into the molecular mechanisms underlying lymphoma pathogenesis and opens a window of opportunity to identify potential diagnostic and prognostic biomarkers.
1. Introduction
Non-Hodgkin lymphomas (NHL) are a very heterogeneous group of lymphoproliferative malignancies characterized by the infiltration of lymphoid tissues
[1]. The majority of NHL are derived from B cells (85% to 90%), while the remaining are derived from T cells or NK cells
[1].
Recently, non-coding RNAs (ncRNAs), which were once thought to be “junk RNA”, have emerged as essential players in the molecular events of normal B-cell development and in lymphomagenesis
[2]. MicroRNAs (miRNAs) are undoubtedly the class of ncRNAs most studied over the years, especially due to their relevant biological function in gene regulation
[3]. MiRNAs are characterized as small ncRNAs with ~22 nucleotides in length, present in all eukaryotic cells, and highly conserved. They function as gene regulators at a post-transcriptional level through binding to the 3′ untranslated region (UTR) of a target mRNA, which results in their repression or degradation
[4]. Recently, the role of miRNAs as regulatory players in B-cell lymphomas is being unveiled, and they have been proposed as potential biomarkers for the diagnosis, prognosis, and prediction of therapy response
[5]. To date, it is established that miRNAs can be found in circulation, not only in its cell-free form but also encapsulated in extracellular vesicles (such as exosomes), which permit them to function in a paracrine manner during lymphoma development and progression (reviewed by Fernandes et al.
[6]).
Recent studies have shown that another class of ncRNAs, known as long non-coding RNAs (lncRNAs), are also master regulators of multiple protein-coding genes and are involved in all cancer hallmarks
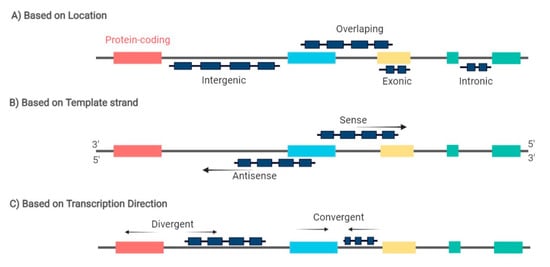
[7][8]. LncRNAs are characterized for being more than 200 nucleotides long and can be further classified based on their biogenesis loci in intronic, exonic, intergenic, or overlapping sense/antisense lncRNAs, and divergent/convergent lncRNAs (
Figure 1)
[9]. These molecules exhibit relatively low expression but high tissue and disease-specific expression patterns
[10]. Among the different functions of lncRNAs in gene expression regulation is the remarkable interplay between lncRNAs and miRNAs, which has the ability to balance miRNA function as miRNA sponges/decoys, creating a competitive endogenous RNA (ceRNA) network
[11][12]. LncRNAs can sequester miRNAs by presenting biding sequences for miRNAs and impairing their functional interaction with mRNA
[13]. Moreover, one lncRNA has the ability to sponge various miRNAs through different biding sites, as seen, for example, for lncRNA MALAT1, which was demonstrated to target miR-101, miR-129, and miR-199a
[14][15][16][17]. Therefore, the miRNA regulatory network is more intricate than previously thought by adding another regulatory layer to the network involving lncRNAs. Recent studies have shown that lncRNAs regulate cell differentiation, and their deregulation plays a key role in the pathogenesis of cancer
[18][19]. In fact, some studies have been analyzing the expression pattern of lncRNAs in the different B-cell lymphoma subtypes. However, compared to solid tumors, there is still a limited number of studies analyzing the role of lncRNAs during normal B-cell development and as key players in B-cell malignancies.
Figure 1. LncRNA can be classified based on: (A) the genomic location between two coding genes in: intronic, exonic, intergenic, and overlapping lncRNA; (B) the template strand from which they are transcribed in: sense and antisense lncRNA; and (C) the direction of lncRNA transcription in: divergent and convergentlncRNA. Arrows indicate the transcription direction. Red, blue, yellow, and green boxes represent exons from different coding genes.
2. MiRNAs and lncRNAs Deregulation in Lymphomagenesis
2.1. The Role of miRNAs in B-Cell Lymphomagenesis
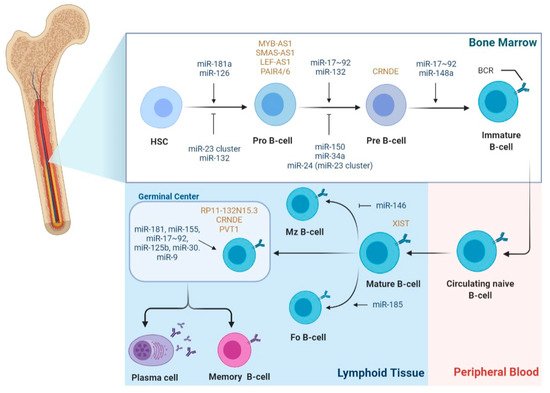
Considering that B-cell development is a highly regulated process, it is not surprising that miRNAs have been implicated in the regulation of most of the stages comprising this process (
Figure 2). Interestingly, during B-cell development, most miRNAs show a stage-specific expression pattern, highlighting their stage-specific function
[20]. The process involving B-cell differentiation seems to be prone to malignant transformation, with increasing evidence showing that disruption of the miRNA network takes part in the initiation and maintenance of lymphomagenesis.
Figure 2. miRNA and lncRNA expression during the different stages of B-cell development. During B-cell development, miRNAs and lncRNAs show a stage-specific expression pattern. For example, miR-181a-5p, miR-150-5p, miR-132-3p, and miR-126-3p were shown to be differentially expressed during the development stages of B cells; in particular, miR-181a-5p ectopic overexpression in common lymphoid progenitors results in an increasing total number of B cells. Conversely, overexpression of miR-23a-5p in HSCs results in the inhibition of B-cell development. MiRNAs are involved in the modulation of the checkpoint of pro to pre-B-cell transition. MiR-132-3p shows a stage-specific and BCR-dependent expression, being normally expressed after the pro-B stage; miR-24-3p, miR-34a, and miR-150-5p, when overexpressed, block the transition at pro to pre-B-cell. In secondary lymphoid tissues, miR-155 and miR-181b are highly expressed in activated B-cells in germinal centers. miR-155 and miR-181b-deficient B cells have defective antibody class switching and differentiation into plasma cells; both miRNAs target activation-induced cytidine deaminase (AID) and PU.1, which promote antibody class switching and antibody production. Other miRNAs, e.g., miR-9, miR-125b, and the miR-30 family, are expressed in GC B cells and enhance plasma-cell differentiation. Concerning lncRNAs regulation of B-cell development, lncRNAs MYB-AS1, SMAS-AS1, and LEF-AS1 were found to play a role in early B cells; CRNDE is overexpressed during proliferating stages, such as pre-B-cells and centroblasts in the GC. LncRNA XIST modulates the X-linked gene regulation from antigen naïve B-cells to activated B-cells during B-cell stimulation. Expression of lncRNAs PVT1 and RP11-132N15.3 were associated with the expression of AID in the GC. (Abbreviations: B-cell receptor (BCR); Follicular B cells (FO B-cells); Hematopoietic stem cells (HSCs); Marginal zone B-cells (MZ B-cells)).
2.2. The Role of LncRNAs in B-Cell Lymphomagenesis
Regarding the regulatory role of lncRNA during the different stages of B-cell development and as drivers of B-cell malignancies, there is still scarcer information when compared to miRNAs. LncRNA expression profiling studies have reported that lncRNA exhibits cell-type-specific expression patterns during the different stages of B-cell differentiation (
Figure 2)
[21][22][23][24]. Consequently, each B-cell subset can be differentiated using its unique lncRNA expression profile
[24]. Petri et al., using a guilt-by-association method, analyzed lncRNAs originated from protein-coding genes with known functions in B-cell development and identified antisense lncRNAs, such as MYB-AS1, SMAS-AS1, and LEF-AS1, with roles in early B cells, associated with RAG2, VPREB1, DNTT, LEF1, SMAD1, and MYB expression
[23]. On the other hand, lncRNA colorectal neoplasia differentially expressed (CRNDE) showed high expression during the proliferating stages, such as pre-B cells and centroblasts in the GC, which was tightly associated with the expression of mitotic cell cycle genes
[23]. In fact, CRNDE was previously demonstrated to be linked to cell-cycle and metabolic changes during proliferation
[25][26]. Brazão et al. identified the expression of PVT1 and some uncharacterized lincRNAs, such as LINC00487, LINC00877, and RP11-132N15.3, associated with the expression of AID and SERPINA9, both specifically expressed in GC centroblasts and centrocytes. Of note, RP11-132N15.3 is described to be encoded approximately 240 kilobases upstream of BCL6
[22][23]. Additionally, based on mice models, several lncRNAs demonstrated a PAX5-dependent expression, a transcription factor involved in B-cell commitment, which were shown to be bound by PAX5 and to have human orthologs previously described
[22].
Non-coding antisense transcripts of PU.1 were reported to inhibit the expression of PU.1 at a translation level, which could indicate its pivotal role in lymphomagenesis given the regulatory function of PU.1 in B-cell differentiation
[27][28].
On the other hand, over the past few years, some studies have been trying to unveil the mechanistic pathways associated with the deregulation of lncRNAs during lymphomagenesis. In this instance, TP53 has been linked to the expression of some lncRNAs in different lymphoma subtypes. In fact, Blume et al. demonstrated, for the first time, the association of lncRNAs and the p53 pathway in CLL and lymphoma by inducing a p53-dependent DNA damage response, which resulted in increased expression of two lncRNAs, NEAT1 and lincRNA-p21, regulating apoptosis or cell-cycle arrest and DNA repair
[29]. In DLBCL, p53 can directly bind to the promoter region of the lncRNA PANDA, which inactivates the MAPK/ERK signaling pathway, suppressing the proliferation of DLBCL cells by a G0/G1 cell-cycle arrest
[30]. Peng et al. demonstrated that lncRNA HULC regulates DLBCL cell apoptosis and cell proliferation via the upregulation of antiapoptotic BCL2 protein and cyclin D1
[31]. The Wnt/β-catenin signaling pathway was shown to be activated by lncRNA FIRRE through promoting the nuclear translocation of β-catenin
[32]. LncRNA DBH-AS1, found to be upregulated in DLBCL, was identified as a positive regulator of cell proliferation, migration, and invasion via binding to the RNA-binding protein BUD13 homolog (BUD13), which in turn regulates fibronectin 1 expression
[33].
2.3. The Role of LncRNAs as ceRNAs in B-Cell Lymphomagenesis
Karreth et al. demonstrated that lncRNA BRAFP1, which is aberrantly expressed in B-cell lymphomas, acts as a ceRNA with BRAF mRNA, increasing its stability and BRAF levels by sequestering specific BRAF-targeting miRNAs, such as miR-134, miR-543, and miR-653. Consequently, BRAF activates MAPK signaling, resulting in DLBCL cells’ proliferation [34]. In fact, NEAT1 was identified as an MYC-regulated transcript promoting DLBCL cells proliferation and lymphomagenesis by regulating the miR-34b-5p-GLI1 pathway [35]. Interestingly, NEAT1, along with LincRNA-p21, were also identified as p53-dependent DNA damage response machinery in lymphoma and CLL [29].
Another reported upregulated lncRNA in DLBCL is MALAT1, whose ceRNA function is through sponging miR-195, resulting in the activation of the immune checkpoint molecule PD-L1 and consequently promoting cell proliferation, migration, and immune escape. Moreover, MALAT1 can induce CD8+ T cell apoptosis and epithelial–mesenchymal transition (EMT)-like processes by regulating the Ras/ERK signaling pathway [36]. In MCL, the knockdown of MALAT1 resulted in cell-cycle arrest and impaired proliferation due to the upregulation of p21 and p27 by EZH2 [37]. MiR-423-5p was reported to be involved in a ceRNA network with lncRNA FOXP4-AS1 in MCL cells. Mechanistically, FOXP4-AS1 acts as a sponge to miR-423-5p, upregulating the expression of NACC1, which results in MCL cell proliferation, migration, and invasion [38].
3. MiRNAs and lncRNAs as Potential Biomarkers for NHL
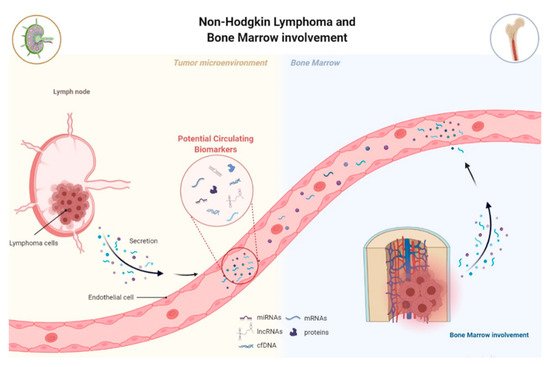
The presence of circulating tumor-associated components, known as “tumor circulome”, which can be easily assessed, appears as a potential option as cancer biomarkers for liquid biopsies (
Figure 3)
[39]. One of the major components of “tumor circulome”, highly present in circulation, are the miRNAs
[40]. MiRNAs emerged as excellent biomarker candidates due to their high stability in biological samples and their high specificity and sensitivity (
Table 1)
[5]. Similarly, increasing evidence has proposed lncRNAs are promising cancer diagnostic and prognostic biomarkers, especially given their high cell type, tissue, and disease type-specific expression (
Table 2). Moreover, lncRNAs have been considered stable and can also be detected in circulation
[41]. However, the majority of studies analyzing deregulated lncRNAs in lymphoma have been performed on tissue samples and cell lines.
Figure 3. Lymphoma-related circulating-free DNA, RNA, or proteins are released by lymphoma cells into circulation, known as tumor circuloma. Analysis of the tumor circuloma can provide a non-invasive approach to screen, diagnose, and surveillance patients during the course of the disease. Moreover, the use of liquid biopsy in substitution of BM biopsy to detect BM infiltration by lymphoma opens the need to readdress the value of the routine standard analysis.
Table 1. Circulating miRNAs as Potential Diagnostic and Prognostic Biomarkers of B-NHL.
| NHL |
miRNA |
Expression |
Biomarker Utility |
Source
Material |
Refs. |
| DLBCL |
miR-155 |
Upregulated |
Diagnostic |
Serum |
[42][43] |
| Subclassification |
Serum |
[44] |
| Prognostic of OS, PFS and RFS |
Plasma,
serum |
[45][46] |
| miR-210 |
Upregulated |
Diagnostic |
Serum |
[42][47] |
| let-7b/c |
Upregulated |
Diagnostic |
Serum |
[48] |
| miR-15a |
Upregulated |
Diagnostic |
Serum |
[43][47][48] |
| miR-16-1 |
Upregulated |
Diagnostic |
Serum |
[43] |
| miR-18a |
Upregulated |
Diagnostic |
Serum |
[48] |
| miR-20a/b |
Upregulated |
Prognostic of OS |
Serum |
[49] |
| miR-21 |
Upregulated |
Diagnostic |
Serum |
[42][47] |
| Subclassification |
Serum |
[50] |
| Monitoring |
Plasma |
[51] |
| Prognostic of OS, PFS and RFS |
Serum |
[42][46][50][52] |
| miR-22 |
Upregulated |
Prognostic of PFS |
Serum |
[53] |
| miR-24 |
Upregulated |
Diagnostic |
Serum |
[48] |
| miR-28 |
Downregulated |
Prognostic of OS, PFS and RFS |
Serum |
[46] |
| miR-29c |
Upregulated |
Diagnostic |
Serum |
[43] |
| miR-33a |
Downregulated |
Prognostic of RFS |
Serum |
[54] |
| miR-34 |
Downregulated |
Diagnostic |
Serum |
[43] |
| miR-92a |
Downregulated |
Diagnostic |
Plasma |
[55] |
| Monitoring |
| miR-93 |
Upregulated |
Prognostic of OS |
Serum |
[49] |
| miR-106a/b |
Upregulated |
Prognostic of OS |
Serum |
[49] |
| miR-125b |
Upregulation |
Prognostic of OS |
Serum |
[56] |
| miR-130a |
Upregulated |
Monitoring |
Serum |
[56] |
| miR-130b |
upregulation |
Prognostic of OS, PFS and RFS |
Serum |
[46] |
| miR-181-5p |
Downregulated |
Subclassification |
Serum |
[44] |
| miR-199-5p |
Upregulated |
Prognostic of OS |
Plasma |
[45] |
| miR-224 |
Upregulated |
Prognostic of RFS |
Serum |
[54] |
| miR-323b |
Downregulated |
Diagnostic |
Serum |
[57] |
| miR-326 |
Upregulated |
Diagnostic |
Serum |
[58] |
| miR-375 |
Downregulated |
Diagnostic |
Serum |
[58] |
| miR-431 |
Downregulated |
Diagnostic |
Serum |
[57] |
| miR-455-3p |
downregulated |
Prognostic of RFS |
Serum |
[54] |
| miR-494 |
upregulated |
Monitoring |
Plasma |
[51] |
| miR-520d-3p |
Upregulated |
Prognostic of RFS |
Serum |
[54] |
| miR-1236 |
Upregulated |
Prognostic of RFS |
Serum |
[54] |
| CLL |
miR-34a |
Upregulated |
Diagnostic |
Serum |
[59] |
| miR-31-5p |
Upregulated |
Diagnostic |
Serum |
[59] |
| miR-150-5p |
Upregulated |
Diagnostic |
Serum |
[59] |
| miR-155-5p |
Upregulated |
Diagnostic |
Serum |
[59] |
| miR-15a-3p |
Upregulated |
Diagnostic |
Serum |
[59] |
| miR-29a-3p |
Upregulated |
Diagnostic |
Serum |
[59] |
Abbreviations: CLL—Chronic lymphocytic leukemia; DLBCL—Diffuse large B-cell lymphoma; OS—Overall Survival; PFS—Progression-free Survival; RFS—Relapse-free Survival.
Table 2. LncRNAs as Potential Diagnostic and Prognostic Biomarkers of NHL.
| NHL |
LncRNA |
Expression |
Biomarker Utility |
Source
Material |
Refs. |
| DLBCL |
PEG10 |
Upregulated |
Diagnostic |
Tissue
Cell lines |
[60] |
| Prognostic of OS |
| LUNAR1 |
Upregulated |
Diagnostic |
Tissue
Cell lines |
[61] |
| Prognostic of OS and PFS |
| FIRRE |
Upregulated |
Diagnostic |
Tissue
Cell lines |
[32] |
| Prognostic of OS |
| HULC |
Upregulated |
Diagnostic |
Tissue
Cell lines |
[31] |
| Prognostic of OS and PFS |
| LINC01857 |
Upregulated |
Diagnostic |
Tissue
Cell lines |
[62] |
| OR3A4 |
Upregulated |
Diagnostic |
Tissue
Cell lines |
[63] |
| Prognostic of OS |
| ENST00000424690 |
Upregulated |
Diagnostic |
Tissue
Cell lines |
[64] |
| ENST00000425358 |
| NR_026892 |
| ENST00000464929 |
Downregulated |
| ENST00000475089 |
| SubSigLnc-17 |
- |
Diagnostic |
Tissue
Cell lines |
[65] |
| Subclassification |
| Prognostic of OS and PFS |
| NONHSAG026900 |
Upregulated |
Diagnostic |
Tissue
Cell lines |
[66] |
| Prognostic of OS and PFS |
| NEAT1_1 |
Upregulated |
Diagnostic |
Tissue
Cell lines |
[67] |
| Prognostic of OS |
| GAS5 |
Upregulated |
Diagnostic |
Tissue
Cell lines |
[68] |
| MIR17HG |
Upregulated |
Diagnostic |
Tissue
Cell lines |
| HULC |
Upregulation |
Diagnostic |
Tissue
Cell lines |
| PCA3 |
Upregulated |
Diagnostic |
Tissue
Cell lines |
| PANDA |
Downregulation |
Diagnostic |
Plasma
Tissue |
[30] |
| Prognostic of OS and RFS |
| TUG1 |
Upregulated |
Diagnostic |
Plasma |
[30] |
| HOTAIR |
Upregulated |
Diagnostic |
Plasma
Tissue |
[69][70] |
Predictive of Treatment
response |
Prognostic of
OS |
| XIST |
Upregulated |
Diagnostic |
Plasma |
[70] |
| GAS5 |
Downregulated |
Diagnostic |
Plasma |
Predictive of Treatment
response |
| 6-lncRNA signature |
- |
Prognostic of OS |
Tissue |
[71] |
| FL |
RP11-625 L16.3 |
Upregulated |
Diagnostic |
Tissue |
[72] |
| RP4-694A7.2 |
Upregulated |
Diagnostic and subclassification |
Tissue |
[73] |
| MCL |
LINK-A |
Upregulated |
Diagnostic |
Plasma |
[74] |
| GATA6-AS |
Downregulated |
Diagnostic |
Plasma |
| MALAT1 |
Upregulated |
Prognostic of OS and DFS |
Tissue
Cell lines |
[37] |
| FOXP4-AS1 |
Upregulated |
Prognostic of OS and DFS |
Plasma |
[38] |
| MORT |
Downregulated |
Diagnostic |
Plasma |
[75] |
| CLL |
lincRNA-p21 |
Downregulated |
Diagnostic |
Plasma |
[76] |
| MM |
TUG1 |
Upregulated |
Diagnostic |
Plasma |
| MALAT1 |
Downregulated |
Diagnostic |
Plasma |
| HOTAIR |
Diagnostic |
Plasma |
| GAS5 |
Diagnostic |
Plasma |
Abbreviations: CLL—Chronic lymphocytic leukemia; DFS—Disease-free Survival; DLBCL—Diffuse large B-cell lymphoma; FL—Follicular lymphoma; MCL—Mantle cell lymphoma; MM—Multiple Myeloma; OS—Overall Survival; PFS—Progression-free Survival; RFS—Relapse-free Survival.
3.1. MiRNAs and lncRNAs as Non-Invasive Diagnostic Biomarkers
A current clinical problem in the context of NHL remains the late and imprecise diagnosis, which in turn negatively conditions patient morbidity and mortality. Several authors have proposed and showed the potential of both individual miRNAs and lncRNAs or panels of these ncRNAs, as biomarkers for diagnosis and subclassification of NHL. Regarding the diagnostic potential, studies have analyzed the expression of miRNAs by comparing NHL cases to healthy controls in order to identify a specific miRNA signature. Lawrie et al.’s study showed, for the first time, that tumor-associated miRNAs, miR-155, miR-210, and miR-21 expression levels were upregulated in DLBCL patients’ serum compared to healthy controls
[42]. Beheshti et al. first established a miRNA expression profile associated with DLBCL development based on cell lines and patient-derived xenograft models, which was then validated in patients’ serum. In the validation study, their profile was reduced to five miRNAs, let-7b, let-7c, miR-18a, miR-24, and miR-15a, which had the ability to discriminate DLBCL patients from healthy donors, with an accuracy of 91%
[48][77].
Circulating miRNAs have also been studied as refiners of the World Health Organization (WHO) classification, leading the way towards a molecular classification of NHL. In fact, the current system of differential diagnosis between the different types of B-cell NHL remains ineffective. For example, Chen et al. demonstrated that plasma levels of miR-21 were higher in DLBCL activated B-cell-like (ABC) compared to the germinal center B-cell-like (GCB), while Bedewy et al. reported higher expression of miR-155 in non-GCB compared to the GCB subtype
[44][50].
Concerning the deregulated expression of lncRNAs in NHL, the vast majority of studies were performed on tissue samples; however, the found tissue-derived deregulated lncRNAs could represent promising circulating biomarkers to be analyzed in future studies. Verma et al. performed a large RNA-seq study of 116 DLBCL tissue samples and identified 2,632 novel lncRNAs, two-thirds of which were only expressed in tumor cells. Moreover, more than one-third of these lncRNAs were found to be differently expressed between ABC and GCB subtypes
[78]. LncRNAs PEG10 and LUNAR1 were found to be upregulated in DBCL patients compared to healthy individuals, with areas under the ROC curve (AUC) up to 0.8228 and 0.9420, respectively, indicating their potential diagnostic value
[61][60].
Regarding circulating NHL-associated lncRNAs, there are still few studies, even though their diagnostic and prognostic potential hs been shown in several solid tumors
[79][80]. The first study on circulating lncRNAs in B-cell lymphoma was performed by Isin et al., which showed downregulation of lincRNA-p21 in CLL patients, upregulation of TUG1 and downregulation of MALAT1, HOTAIR, and GAS5 in MM patients
[76].
3.2. MiRNAs and lncRNAs as Prognostic Biomarkers
Several studies have been exploring the value of circulating miRNAs as prognostic markers for NHL. In 2008, Lawrie et al. were the first to report that high serum levels of miR-21 were associated with relapse-free survival (RFS) in DLBCL patients, which were later on supported in other studies [42][50]. Similarly, high serum levels of miR-22 at diagnosis in DLBCL were associated with a worse progression-free survival (PFS), independently of the currently used clinical prognostic index [53]. The upregulation of circulating miR-155 and miR-125b was associated with shorter overall survival (OS) of DLBCL patients, while miR-20a/b, miR-93, and miR-106a/b plasma profiles were associated with higher mortality in DLBCL [45][49][56]. Song et al.’s study reported that elevated levels of miR-224, miR-520d-3p, and miR-1236 and lower levels of miR-33a and miR-455-3p were associated with lower medium remission time, and consequently, higher probability of remission, independently of IPI score [54]. A 4-miRNA expression profile (higher levels of miR-21, miR-130b, miR-155; lower levels of miR-28) was shown to be associated with relapse, as well as inferior PFS and OS after R-CHOP, independently of IPI score [46].
he 6-lncRNA signature defined by Sun et al. was shown to be associated with patients OS, independently of standard clinical factors, and permitted to stratify DLBCL patients in high and low-risk groups, improving survival prediction [71]. Moreover, the SubSigLnc-17 profile was not only able to discriminate clinically molecular DLBCL subtypes but also was shown to be significantly associated with patients’ OS and PFS [65]. The expression levels of lncRNA HOTAIR were not only associated with tumor size and clinical stage but also with the presence of B symptoms and IPI score. In fact, higher levels of HOTAIR were associated with better patients’ prognoses, being characterized as an independent predictive biomarker for DLBCL [69][70].
The assessment of patients’ prognosis and decisions on treatment alteration are mostly based on imaging PET-CT and clinical evaluations [81]. However, due to the insufficient sensitivity and specificity of these tools, there is an impending need to identify new predictors that permit the early identification of patients with inherent or acquired the refractory disease during treatment. Detection of miRNAs or lncRNAs can lower the detection limit of disease beyond the capabilities of current methods and create a “window of opportunity” for intervention prior to clinical relapse. Earlier initiation of second-line therapy at a point of minimal tumor burden may improve patients’ outcomes. Song et al. identified a 5-miRNA profile (miR-224, miR-455-3p, miR-1236, miR-33a, and miR-520d-3p) associated with R-CHOP response in DLBCL patients, being a significant predictor of response, independent from the IPI score [54].
Regarding lncRNAs as treatment response biomarkers, Senousy et al. observed that pretreatment circulating levels of HOTAIR were higher, whereas GAS5 were lower in non-responders compared to responders to R-CHOP. Moreover, when performing multivariate analysis, HOTAIR appeared as an independent predictor of R-CHOP failure [70]. DLBCL patients with higher expression levels of NONHSAG026900 were shown to have a better response to chemotherapy compared to patients with lower levels [66].
3.3. Clinical Trials for Potential miRNA and lncRNA Biomarkers
Since 2008, when Lawrie et al. demonstrated for the first time the potential of miRNAs as biomarkers in DLBCL patients, a considerable number of clinical trials have been registered to clinically validate them
[42]. A clinical trial (NCT01505699) involving 186 patients with B-cell acute lymphoblastic leukemia analyzed microRNA signature levels and their association with different clinical outcomes
[82]. On the other hand, clinical trial NCT01057199 specifically tested miR-34a and miR-194 as biomarkers in cell samples from patients with acute myeloid leukemia
[83]. Moreover, clinical trial NCT01541800 especially focused on pediatric cancers (central nervous system tumors, leukemia, and lymphoma), evaluated the presence of circulating miRNAs in the blood and cerebrospinal fluid of patients under chemotherapy
[84].
Currently, there is no clinical trial exploring the possible application of lncRNAs as lymphoma biomarkers; however, there are some lncRNAs undergoing clinical trials or being patented for solid tumors, such as lung, colorectal, thyroid, breast, and gynecologic cancers
[85][86]. To date, the lncRNA PCA3 was the first and only lncRNA approved for clinical practice as an early diagnostic biomarker for prostate cancer
[87]. There is only one registered clinical trial, currently active, for hematological cancer to study the correlation between lncRNA XIST and immunophenotyping AML patients (NCT04288739)
[88]. Moreover, few clinical trials have been exploring the possible application of combined ncRNA profiles, especially miRNAs and lncRNAs, as biomarkers for the diagnosis and prognosis of cancer
[86].