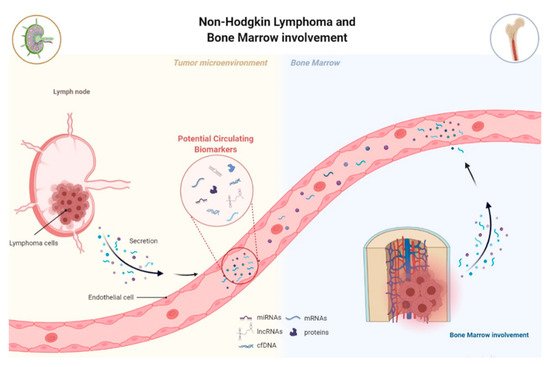
Non-Hodgkin lymphoma (NHL) is a heterogeneous malignancy with variable patient outcomes. There is still a lack of understanding about the different players involved in lymphomagenesis, and the identification of new diagnostic and prognostic biomarkers is urgent. MicroRNAs and long non-coding RNAs emerged as master regulators of B-cell development, and their deregulation has been associated with the initiation and progression of lymphomagenesis. They can function by acting alone or, as recently proposed, by creating competing endogenous RNA (ceRNA) networks. The study of miRNAs’ and lncRNAs’ deregulation in NHL, either alone or as ceRNAs networks, offers new insights into the molecular mechanisms underlying lymphoma pathogenesis and opens a window of opportunity to identify potential diagnostic and prognostic biomarkers.
- lymphoma
- non-Hodgkin’s lymphoma
- miRNAs
- lncRNAs
- biomarkers
- ceRNA network
1. Introduction
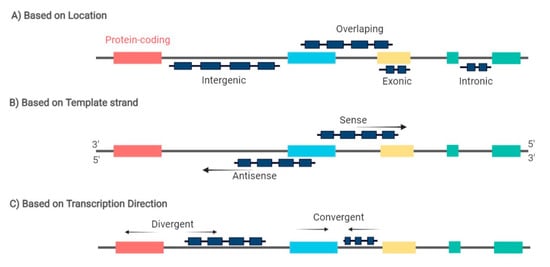
2. MiRNAs and lncRNAs Deregulation in Lymphomagenesis
2.1. The Role of miRNAs in B-Cell Lymphomagenesis
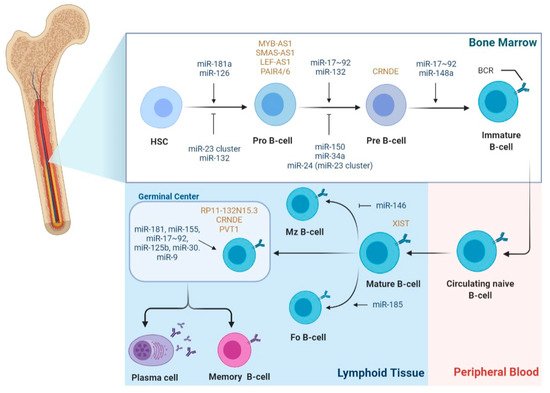
2.2. The Role of LncRNAs in B-Cell Lymphomagenesis
2.3. The Role of LncRNAs as ceRNAs in B-Cell Lymphomagenesis
Karreth et al. demonstrated that lncRNA BRAFP1, which is aberrantly expressed in B-cell lymphomas, acts as a ceRNA with BRAF mRNA, increasing its stability and BRAF levels by sequestering specific BRAF-targeting miRNAs, such as miR-134, miR-543, and miR-653. Consequently, BRAF activates MAPK signaling, resulting in DLBCL cells’ proliferation [87]. In fact, NEAT1 was identified as an MYC-regulated transcript promoting DLBCL cells proliferation and lymphomagenesis by regulating the miR-34b-5p-GLI1 pathway [88]. Interestingly, NEAT1, along with LincRNA-p21, were also identified as p53-dependent DNA damage response machinery in lymphoma and CLL [74].
Another reported upregulated lncRNA in DLBCL is MALAT1, whose ceRNA function is through sponging miR-195, resulting in the activation of the immune checkpoint molecule PD-L1 and consequently promoting cell proliferation, migration, and immune escape. Moreover, MALAT1 can induce CD8+ T cell apoptosis and epithelial–mesenchymal transition (EMT)-like processes by regulating the Ras/ERK signaling pathway [92]. In MCL, the knockdown of MALAT1 resulted in cell-cycle arrest and impaired proliferation due to the upregulation of p21 and p27 by EZH2 [83]. MiR-423-5p was reported to be involved in a ceRNA network with lncRNA FOXP4-AS1 in MCL cells. Mechanistically, FOXP4-AS1 acts as a sponge to miR-423-5p, upregulating the expression of NACC1, which results in MCL cell proliferation, migration, and invasion [93].
3. MiRNAs and lncRNAs as Potential Biomarkers for NHL
| NHL | miRNA | Expression | Biomarker Utility | Source Material |
Refs. |
|---|---|---|---|---|---|
| DLBCL | miR-155 | Upregulated | Diagnostic | Serum | [110,111] |
| Subclassification | Serum | [112] | |||
| Prognostic of OS, PFS and RFS | Plasma, serum |
[113,114] | |||
| miR-210 | Upregulated | Diagnostic | Serum | [110,115] | |
| let-7b/c | Upregulated | Diagnostic | Serum | [116] | |
| miR-15a | Upregulated | Diagnostic | Serum | [111,115,116] | |
| miR-16-1 | Upregulated | Diagnostic | Serum | [111] | |
| miR-18a | Upregulated | Diagnostic | Serum | [116] | |
| miR-20a/b | Upregulated | Prognostic of OS | Serum | [117] | |
| miR-21 | Upregulated | Diagnostic | Serum | [110,115] | |
| Subclassification | Serum | [118] | |||
| Monitoring | Plasma | [119] | |||
| Prognostic of OS, PFS and RFS | Serum | [110,114,118,120] | |||
| miR-22 | Upregulated | Prognostic of PFS | Serum | [121] | |
| miR-24 | Upregulated | Diagnostic | Serum | [116] | |
| miR-28 | Downregulated | Prognostic of OS, PFS and RFS | Serum | [114] | |
| miR-29c | Upregulated | Diagnostic | Serum | [111] | |
| miR-33a | Downregulated | Prognostic of RFS | Serum | [122] | |
| miR-34 | Downregulated | Diagnostic | Serum | [111] | |
| miR-92a | Downregulated | Diagnostic | Plasma | [123] | |
| Monitoring | |||||
| miR-93 | Upregulated | Prognostic of OS | Serum | [117] | |
| miR-106a/b | Upregulated | Prognostic of OS | Serum | [117] | |
| miR-125b | Upregulation | Prognostic of OS | Serum | [124] | |
| miR-130a | Upregulated | Monitoring | Serum | [124] | |
| miR-130b | upregulation | Prognostic of OS, PFS and RFS | Serum | [114] | |
| miR-181-5p | Downregulated | Subclassification | Serum | [112] | |
| miR-199-5p | Upregulated | Prognostic of OS | Plasma | [113] | |
| miR-224 | Upregulated | Prognostic of RFS | Serum | [122] | |
| miR-323b | Downregulated | Diagnostic | Serum | [125] | |
| miR-326 | Upregulated | Diagnostic | Serum | [126] | |
| miR-375 | Downregulated | Diagnostic | Serum | [126] | |
| miR-431 | Downregulated | Diagnostic | Serum | [125] | |
| miR-455-3p | downregulated | Prognostic of RFS | Serum | [122] | |
| miR-494 | upregulated | Monitoring | Plasma | [119] | |
| miR-520d-3p | Upregulated | Prognostic of RFS | Serum | [122] | |
| miR-1236 | Upregulated | Prognostic of RFS | Serum | [122] | |
| CLL | miR-34a | Upregulated | Diagnostic | Serum | [127] |
| miR-31-5p | Upregulated | Diagnostic | Serum | [127] | |
| miR-150-5p | Upregulated | Diagnostic | Serum | [127] | |
| miR-155-5p | Upregulated | Diagnostic | Serum | [127] | |
| miR-15a-3p | Upregulated | Diagnostic | Serum | [127] | |
| miR-29a-3p | Upregulated | Diagnostic | Serum | [127] |
| NHL | LncRNA | Expression | Biomarker Utility | Source Material |
Refs. |
|---|---|---|---|---|---|
| DLBCL | PEG10 | Upregulated | Diagnostic | Tissue Cell lines |
[128] |
| Prognostic of OS | |||||
| LUNAR1 | Upregulated | Diagnostic | Tissue Cell lines |
[86] | |
| Prognostic of OS and PFS | |||||
| FIRRE | Upregulated | Diagnostic | Tissue Cell lines |
[77] | |
| Prognostic of OS | |||||
| HULC | Upregulated | Diagnostic | Tissue Cell lines |
[76] | |
| Prognostic of OS and PFS | |||||
| LINC01857 | Upregulated | Diagnostic | Tissue Cell lines |
[94] | |
| OR3A4 | Upregulated | Diagnostic | Tissue Cell lines |
[129] | |
| Prognostic of OS | |||||
| ENST00000424690 | Upregulated | Diagnostic | Tissue Cell lines |
[130] | |
| ENST00000425358 | |||||
| NR_026892 | |||||
| ENST00000464929 | Downregulated | ||||
| ENST00000475089 | |||||
| SubSigLnc-17 | - | Diagnostic | Tissue Cell lines |
[131] | |
| Subclassification | |||||
| Prognostic of OS and PFS | |||||
| NONHSAG026900 | Upregulated | Diagnostic | Tissue Cell lines |
[132] | |
| Prognostic of OS and PFS | |||||
| NEAT1_1 | Upregulated | Diagnostic | Tissue Cell lines |
[133] | |
| Prognostic of OS | |||||
| GAS5 | Upregulated | Diagnostic | Tissue Cell lines |
[134] | |
| MIR17HG | Upregulated | Diagnostic | Tissue Cell lines |
||
| HULC | Upregulation | Diagnostic | Tissue Cell lines |
||
| PCA3 | Upregulated | Diagnostic | Tissue Cell lines |
||
| PANDA | Downregulation | Diagnostic | Plasma Tissue |
[75] | |
| Prognostic of OS and RFS | |||||
| TUG1 | Upregulated | Diagnostic | Plasma | [75] | |
| HOTAIR | Upregulated | Diagnostic | Plasma Tissue |
[80,135] | |
| Predictive of Treatment response |
|||||
| Prognostic of OS |
|||||
| XIST | Upregulated | Diagnostic | Plasma | [135] | |
| GAS5 | Downregulated | Diagnostic | Plasma | ||
| Predictive of Treatment response |
|||||
| 6-lncRNA signature | - | Prognostic of OS | Tissue | [136] | |
| FL | RP11-625 L16.3 | Upregulated | Diagnostic | Tissue | [137] |
| RP4-694A7.2 | Upregulated | Diagnostic and subclassification | Tissue | [138] | |
| MCL | LINK-A | Upregulated | Diagnostic | Plasma | [139] |
| GATA6-AS | Downregulated | Diagnostic | Plasma | ||
| MALAT1 | Upregulated | Prognostic of OS and DFS | Tissue Cell lines |
[83] | |
| FOXP4-AS1 | Upregulated | Prognostic of OS and DFS | Plasma | [93] | |
| MORT | Downregulated | Diagnostic | Plasma | [140] | |
| CLL | lincRNA-p21 | Downregulated | Diagnostic | Plasma | [141] |
| MM | TUG1 | Upregulated | Diagnostic | Plasma | |
| MALAT1 | Downregulated | Diagnostic | Plasma | ||
| HOTAIR | Diagnostic | Plasma | |||
| GAS5 | Diagnostic | Plasma |
3.1. MiRNAs and lncRNAs as Non-Invasive Diagnostic Biomarkers
3.2. MiRNAs and lncRNAs as Prognostic Biomarkers
Several studies have been exploring the value of circulating miRNAs as prognostic markers for NHL. In 2008, Lawrie et al. were the first to report that high serum levels of miR-21 were associated with relapse-free survival (RFS) in DLBCL patients, which were later on supported in other studies [110,118]. Similarly, high serum levels of miR-22 at diagnosis in DLBCL were associated with a worse progression-free survival (PFS), independently of the currently used clinical prognostic index [121]. The upregulation of circulating miR-155 and miR-125b was associated with shorter overall survival (OS) of DLBCL patients, while miR-20a/b, miR-93, and miR-106a/b plasma profiles were associated with higher mortality in DLBCL [113,117,124]. Song et al.’s study reported that elevated levels of miR-224, miR-520d-3p, and miR-1236 and lower levels of miR-33a and miR-455-3p were associated with lower medium remission time, and consequently, higher probability of remission, independently of IPI score [122]. A 4-miRNA expression profile (higher levels of miR-21, miR-130b, miR-155; lower levels of miR-28) was shown to be associated with relapse, as well as inferior PFS and OS after R-CHOP, independently of IPI score [114].
he 6-lncRNA signature defined by Sun et al. was shown to be associated with patients OS, independently of standard clinical factors, and permitted to stratify DLBCL patients in high and low-risk groups, improving survival prediction [136]. Moreover, the SubSigLnc-17 profile was not only able to discriminate clinically molecular DLBCL subtypes but also was shown to be significantly associated with patients’ OS and PFS [131]. The expression levels of lncRNA HOTAIR were not only associated with tumor size and clinical stage but also with the presence of B symptoms and IPI score. In fact, higher levels of HOTAIR were associated with better patients’ prognoses, being characterized as an independent predictive biomarker for DLBCL [80,135].
The assessment of patients’ prognosis and decisions on treatment alteration are mostly based on imaging PET-CT and clinical evaluations [151]. However, due to the insufficient sensitivity and specificity of these tools, there is an impending need to identify new predictors that permit the early identification of patients with inherent or acquired the refractory disease during treatment. Detection of miRNAs or lncRNAs can lower the detection limit of disease beyond the capabilities of current methods and create a “window of opportunity” for intervention prior to clinical relapse. Earlier initiation of second-line therapy at a point of minimal tumor burden may improve patients’ outcomes. Song et al. identified a 5-miRNA profile (miR-224, miR-455-3p, miR-1236, miR-33a, and miR-520d-3p) associated with R-CHOP response in DLBCL patients, being a significant predictor of response, independent from the IPI score [122].
Regarding lncRNAs as treatment response biomarkers, Senousy et al. observed that pretreatment circulating levels of HOTAIR were higher, whereas GAS5 were lower in non-responders compared to responders to R-CHOP. Moreover, when performing multivariate analysis, HOTAIR appeared as an independent predictor of R-CHOP failure [135]. DLBCL patients with higher expression levels of NONHSAG026900 were shown to have a better response to chemotherapy compared to patients with lower levels [132].
3.3. Clinical Trials for Potential miRNA and lncRNA Biomarkers
This entry is adapted from the peer-reviewed paper 10.3390/biomedicines9121934
