Tuberculosis (TB), caused by the opportunistic pathogen
Mycobacterium tuberculosis (
M.tb), is a deadly disease and a major cause of death globally,
[1] The prognosis of TB is further worsened due to co-morbid conditions, such as infections of HIV, and now the ongoing COVID-19 pandemic is posing additional challenges
[2]. The emergence of drug resistant forms (MDR and XDR) of
M.tb is a cause of concern as it has slowed our efforts to eradicate TB worldwide
[3]. Macrophages are primarily efficient in clearing pathogens; however,
M.tb can survive intracellularly within the niche of macrophage itself.
M.tb has evolved various mechanisms that allow it to hijack the process of phagosome-lysosome fusion, inhibit acidification of phagosome, suppress autophagy and apoptosis pathways used by macrophage for the clearance of pathogens
[4][5][6][7].
M.tb overpowers the extremely microbicidal nature presented within the macrophages through a multifaceted and complex interplay between its proteins and host immune responses
[8][9]. Several
M.tb proteins have been reported to evoke innate and adaptive immune responses, though many of these act as decoy antigens to subvert the immune system. Decoy antigens mimic host-pathogen effector components and can misdirect the immune response pathways that favor the survival of the pathogen. Pathogenic bacteria such as
M.tb also use decoy proteins as a generic mechanism to mask themselves from immune surveillance, thereby evading and subverting host immune responses
[10][11]. The decoy antigens can be classified into three broad categories, namely, receptor, bodyguard and sensing decoys. As the name suggests, receptor decoys are employed by the pathogens to modulate host immune signaling pathways. In contrast, bodyguard decoys act as inactive mimics to safeguard the virulence factors of pathogens from the host response. Sensing decoys mimic the effector functions of the target proteins of both the host and the pathogens
[12].
The mycobacterial PE/PPE/PE_PGRS protein family, present only in the genus mycobacterium and nowhere else in the living kingdom, occupies approximately 10% of the coding capacity of the
M.tb genome. Despite the reductive genomic evolution of
M.tb [13], the PE/PPE/PE_PGRS family of genes has been expanding during mycobacterial evolution. The presence of this family only in pathogenic strains of the genus mycobacterium, such as
M.tb, M.marinum and
M.bovis, points to its likely importance in disease pathogenesis
[14]. The evolution of PE/PPE gene families was found to be associated with the ESX secretion system
[15], and proteins were majorly reported to be either surface exposed or secreted
[16][17][18]. Cell-surface localization of PE/PPE/PE_PGRS proteins may serve an important function in host-pathogen interactions and in the virulence and pathogenesis of
M.tb [19]. PE_PGRS (polymorphic GC-rich sequences) proteins are a subclass of the PE protein family and consists of a highly conserved N-terminal (approx. 110 amino acid long) the PE domain followed by the C-terminal domain harboring multiple repeats of Gly-Gly-Ala or Gly-Gly-Asn. Deciphering the role of proteins belonging to the PE_PGRS family may reveal new aspects of the biology of
M.tb. The presence of multiple tandem repeats of GGA or GGN has been attributed to cause antigenic variations and aid in immune evasion mechanisms, thereby facilitating pathogen survival. The repetitive nature of PE_PGRS proteins and their surface localization both lead to the generation of immune responses by macrophages and may aid in immune subversion
[17].
2. Immune Evasion and Subversion Properties of PE_PGRS Proteins: A Possible Reflection of Antigenic Variation, Disordered Nature and Glycine Content
During the course of evolution, pathogenic bacteria developed multiple strategies to avoid or subvert host machinery, especially the mechanisms that drive protective outcomes of host immune response
[20][21]. Pathogens also manipulate the outcome of the host’s immune response by altering antigen presentation pathways and engaging host immune machinery with multiple antigens
[22][23].
M.tb utilize extremely progressive and harmonized mechanisms of immune evasion that divert or subvert the host proteins involved in neutralizing the virulence of the pathogen. In doing so, the host machinery gets engaged in evoking immune responses against the decoy antigens, thereby neutralizing the efficacy of host immune response in bacterial clearance
[10][11]. Multiple PE_PGRS proteins evoke different signals that allow the pathogen to evade the host immune response
[24]. PGRS domain of PE_PGRS62 protects the PE protein from ubiquitin-proteasome mediated degradation and also affects the ability of the CD8
+ T-cells to recognize the protein, thereby conferring protection to the pathogen present within the macrophages
[25].
Several pathogens employ intrinsically disordered proteins (IDPs) or disordered short stretches for a variety of moonlighting functions
[26][27][28]. IDPs, by virtue of their conformational plasticity and short interaction motifs, can interact with different protein partners
[29]. Such disordered effector proteins perturb host cellular cascades via favorable interactions through molecular mimicry in both viruses and bacteria
[27][28][30]. The PGRS domain of PE_PGRS proteins lacks a definite three-dimensional (3D) structure and is intrinsically disordered in nature
[16][31][32][33]. The transition from an ordered to a disordered state or vice versa will serve to hijack host immune machinery for subsequent survival of the pathogen
[13][16][34].
The generation of antigenic variation is one of the passive mechanisms of immune evasion and subversion
[35][36]. PE/PPE/PE_PGRS proteins are known to provide a major source of antigenic variations in
M.tb and its clinical isolates
[17][37]. Thus, their prospective importance in acting as a decoy antigen to the host is emphasized. The interaction of
M.tb with macrophage offsets the Ca
2+ signaling that causes abnormality in phagosome maturation. Ca
2+ binds with the PE_PGRS33 and PE_PGRS61 proteins
[24][38]. These calcium dependent PE_PGRS proteins decrease the Ca
2+ concentration during the initial phase of non-specific attachment of
M.tb with the alveolar macrophages. The decrease in the Ca
2+ in the macrophage suppresses the phagolysosomal fusion of the
M.tb with the acidic lysosome; thereby contributing to the survival of the
M.tb. PE_PGRS 33 and PE_PGRS41 are cell wall associated proteins. While the PE domain of the PE_PGRS 33 is important for cellular localization, the PGRS domain of this protein is important for cellular morphology of the bacterium and its entry within the host cells. Knock-in of the
PE_PGRS33 gene in
M smegmatis imparts endurance to the bacterium to overcome the cytotoxic effect of the macrophage and enhances the level of TNF. Although
M smegmatis does not effectively infect host cells, recombinant strains of
M smegmatis expressing PE_PGRS33 can colonize the lungs, spleen and liver, which is a typical feature for virulent
M tuberculosis [34]. Ramakrishnan et al. showed that pathogenic
M.marinum expresses two proteins (mag 24 and mag 85) that are homologous with
M.tb PE_PGRS protein family, and are involved in granuloma formation and the replication of the pathogen within the macrophage
[39]. Mice immunized with the PE domain of the PE_PGRS 33 exhibit a higher cell-mediated response while immunization with the complete PE_PGRS 33 leads to increased humoral response
[40]. These studies also suggest that differential expression and the regulation of PE/PE_PGRS protein family during
M.tb infection play a key role in enhancing the virulence features of the pathogen.
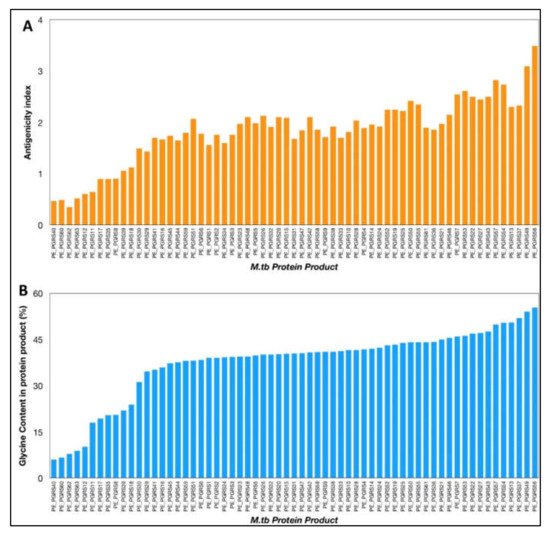
The VaxiJen antigenicity prediction tool shows a high antigenicity index for PE_PGRS proteins (
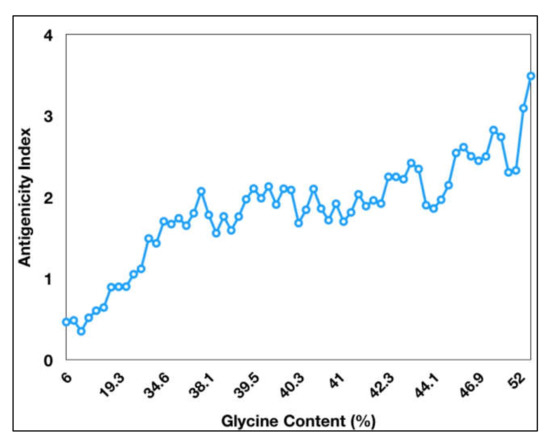
Figure 1). The antigenicity index of PE_PGRS proteins increases as a direct function of the glycine content of these proteins (
Figure 2). PGRS domain of PE_PGRS proteins was observed to be highly rich in glycine, with major chunks of Gly-Gly-Ala stretches similar to EBNA-1 antigen
[25]. Glycine, a highly conserved amino acid, is known to initiate several protective and immunomodulatory responses in the host cells. Glycine modulates the function of the macrophage and evokes inflammatory cytokines, as compared to other amino acids
[41]. Cell wall proteins that are rich in glycine exhibit greater antigenicity and are notable targets in several autoimmune and food borne allergies. It is important to note that the presence of high glycine content in proteins with high antigenicity indices is not just a matter of chance but points to the role of glycine-rich proteins in non-specific but targeted protective immune responses from host macrophages.
Figure 1. (A) Antigenicity index of PE_PGRS proteins of M.tb, as predicted by antigenicity prediction tool VaxiJen. (B) Glycine content of PE_PGRS proteins of M.tb calculated by ExpasyProtParam tool. All values were plotted in increasing order of their magnitude.
Figure 2. Antigenicity index of PE_PGRS proteins increases with increase in glycine content of PE_PGRS proteins. Antigenicity index was plotted against glycine percentage in linear ratio.
The role of PE_PGRS proteins in the immune evasion mechanism is attributed to varied and diverse patterns of the cytokine profile during
M.tb infections
[42]. While some of the PE_PGRS family of proteins, such as PE_PGRS5, PE_PGRS11, PE_PGRS17 and PE_PGRS30, evoke pro-inflammatory responses; others such as PE_PGRS26 are known to induce anti-inflammatory responses. This shows that PE_PGRS have contrasting roles in immune response and can act as a molecular switch for skewing the response as pro-host or pro-pathogen during tuberculosis
[14][37]. The partial homology of PE-PGRS with EBNA domain of the Epstein–Barr virus speculates that it may play a role in the evasion of cytotoxic T-cell response to inhibit antigen processing
[40][43].
Protein antigens are processed through the MHC (major histocompatibility complex) class I and MHC class II. MHC I is ubiquitously expressed on nucleated cells whereas MHC II is expressed on antigen presenting cells (APCs) including macrophages, dendritic cells, etc. Within the macrophage,
M.tb secreted proteins are processed into smaller peptides and presented through the MHC II to the T-cells
[44]. The proteins are processed through the proteasomal degradation machinery of the cell, which are translocated to the endoplasmic reticulum through the transporters associated with antigen processing (TAP) proteins
[45]. CD4
+ T cells recognize these processed antigens primed on the MHC II leading to the generation of effector and memory T-cell response against the antigenic peptides.
M.tb involves multiple mechanisms to prevent or bypass antigen presentation processes (pathways) by inhibiting the truncation of secreted proteins into 8–25 amino acid long short peptides, required for the MHC II pathway
[10][44][46]. Phagosomes, the main component of the MHC class II mediated classical antigen presentation pathway is a critical spot within the macrophages that is hijacked by the
M.tb, resulting in inhibition of the proteasomal processing of secreted antigens. Thus,
M.tb antigens within the macrophage are masked from being recognized by the T-cells, thereby protecting
M.tb from cellular immune response
[47]. PE/PPE/PE_PGRS proteins could be expressed as the early immunodominant antigens followed by the other functionally dominant but immuno-subdominant virulence factors. PE_PGRS proteins neutralize the effector functions of the host immune system, thereby acting as “decoy” for allowing the safe passage of other important effector molecules of the pathogen within the internal proximity of the host. Effector T cells primed against the decoy immunogen search for similar antigens throughout the cells of the host, which are discontinued by the pathogen during a subsequent phase of infection. In this way, the dominant virulent factors of
M.tb remain unaffected by the cell-mediated immune response. The consequent subversion of T-cell response allows the bacteria to successfully establish its pathogenicity and disease progression within the host
[48].
Several members of the PE_PGRS protein family were shown to induce a wide range of contradictory T-cell and B-cell responses as described in earlier sections
[49]. Such responses are not specific to this protein family, rather a generalized and diverse immune profile have been observed
[42]. PE_PGRS11 and PE_PGRS17 proteins are involved in the activation and maturation of human dendritic cells and boost pro-inflammatory responses
[50]. The PE and PGRS domain of PE_PGRS33 evoke different immune response against
M.tb. Mice immunized with PE domain of Rv1818c elicited cellular responses and IFN-γ production, while the humoral response was induced upon immunization with the PGRS domain and not by the PE domain alone
[40]. Another study showed the generation of B-cell responses against the PGRS domain of PE_PGRS33
[51].
M.smegmatis over-expressing PE_PGRS33 and PE_PGRS26 show enhanced production of IL-10 cytokine levels in macrophage cell lines
[52]. An anti-inflammatory response of macrophages due to the PE_PGRS30 protein in terms of the reduced production of IL-12, TNF-α and IL-6 was reported
[53]. PE_PGRS33 is linked with the increased production of TNF-α and IL-10, and reduced levels of IL-12p40
[54]. In contrast, the expression of PE_PGRS16 enhances IL-12p40 levels but reduces IL-10 cytokine production
[52]. The immune response generated by PE_PGRS16 was antagonistic to that of PE_PGRS26
[52][55]. These studies show that the PGRS domain plays a key role in PE_PGRS proteins and is an important target for manipulating immune response.
The elicitation of antibody responses specifically directed against the glycine and asparagine repeats has been reported
[56]. PPE18 and some other 20 PE proteins have been shown to generate CD4 or CD8 mediated T-cell responses
[57]. Th-2 responses and reduced IFN-Υ levels have been detected against PPE44 protein of
M.tb [58]. PGRS domain of PE_PGRS5 protein induce TNF-α and IL-12 cytokines in macrophages
[59] in a calcium dependent manner
[60].
One of the most widely used anti-TB vaccine strains, BCG, is not fully capable of secreting a class of PE/PPE family proteins (specifically PE_PGRS and PPE-MPTR) due to the absence of the RD5-genetic region (containing functional Esx-5 and PPE38/71 involved in secretion)
[61]. The BCG vaccine elicits a reduced repertoire of antigens during infection. In order to assess the immunogenic potential of PE/PPE/PE_PGRS proteins, Ates et al. restored the BCG strain with PPE38 locus, which improved the PE_PGRS and PPE_MPTR secretion in infected mice. Restoration of PE_PGRS and PPE_MPTR secretion neither enhanced the activation of immune cells nor boosted the protective efficacy of the restored BCG mutant strain
[61]. Further studies are warranted to reveal the role of PGRS domain in improving the efficacy of recombinant BCG.
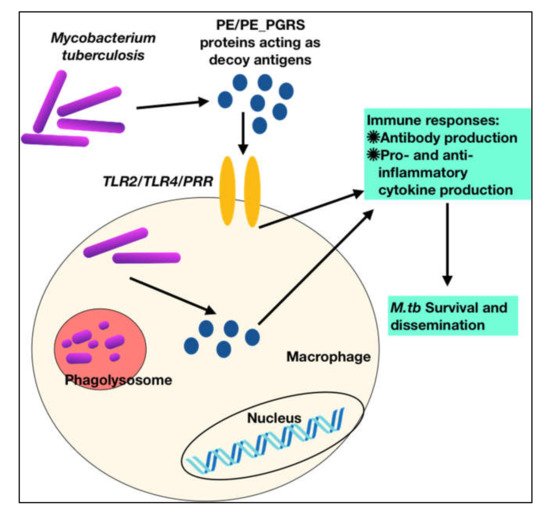
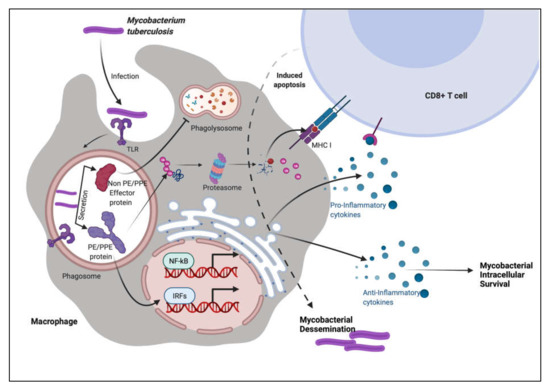
To summarize, these observations show that PE PGRS proteins have a variety of contrasting implications, not simply the PGRS domain, which may aid in evasion and modification of immune effector activities, and hence undermine the targeting of other critical mycobacterial pathogenic proteins (Figure 3 and Figure 4). This subversion may influence the course of disease pathogenesis and lead to higher survival rates of M.tb within alveolar macrophages. These observations are a pointer to reconsider the immunomodulatory effects of PE/PPE/PE_PGRS proteins (Table 1, Table 2, Table 3 and Table 4), few of which are considered in vaccine formulations. Understanding the mechanisms of the PE/PE_PGRS family of proteins in evading and subverting immune responses may aid in targeting these proteins for future therapeutic interventions.
Figure 3. M.tb PPE_PGRS antigens play a role of virulent determinants by acting as an immunological decoy to capture the host immune machinery and evoke varied immune responses. This aids in evasion and subversion of host immune cellular functions during M.tb infection.
Figure 4. PE/PPE proteins augment the immune system of the host using decoy strategies. M.tb infection is most commonly found in macrophages, where the pathogen is endocytosed and transported to the endosome compartment. M.tb secretes non-PE/PPE and PE/PPE proteins along with other effector molecules. PE/PPE proteins are involved in the activation of immune cells. These proteins, according to the immune system, pose the greatest hazard to the cellular system. Other non-PE/PPE effectors, on the other hand, infiltrate the system and take control of the machinery, inflicting severe damage and pathogenicity.
Table 1. Comprehensive table showing role of different PE proteins in immune modulation of host.
| Sr. No. |
PE Proteins |
Role in Immune Modulation |
Reference |
| 1. |
PE17 |
|
[62][63] |
|
|
| 2. |
PE6 |
|
[64][62] |
| 3. |
PE31 |
|
[64][65] |
| 4. |
PE13 |
|
[66] |
| 5. |
PE27 |
|
[67] |
| 6. |
PE11 |
|
[68] |
| 7. |
PE5 |
|
[69] |
| 8. |
PE15 |
|
[69] |
Table 2. Comprehensive table showing role of different PPE proteins in host immune modulation.
| Sr. No. |
PPE Proteins |
Role in Immune Modulation |
Reference |
| 1. |
PPE18 |
|
[70] |
| 2. |
PPE65 |
|
[71] |
| 3. |
PPE57 |
|
[72] |
| 4. |
PPE26 |
|
[73] |
| 5. |
PPE60 |
-
Initiates macrophage pyroptosis via caspases/NLRP3/gasdermin
-
Pro-inflammatory cytokines are stimulated
-
TLR-2 agonist
-
Activates Th-1/Th-17 responses in macrophages
|
[74][75] |
| 6. |
PPE11 |
|
[76] |
| 7. |
PPE27 |
|
[77] |
| 8. |
PPE44 |
|
[78] |
| |
|
|
|
| 9. |
PPE38 |
|
[79] |
| 10. |
PPE10 |
|
[80] |
| 11. |
PPE32 |
-
Through ERK1/2 signaling, it boosts the expression of IL-12p40 and IL-32
-
Promotes macrophage apoptosis
|
[81] |
| 12. |
PPE57 |
|
[63] |
Table 3. Comprehensive table showing role of different PE-PGRS proteins in host immune modulation.
| Sr. No. |
PE_PGRS Proteins |
Role in Immune Modulation |
Reference |
| 1. |
PE_PGRS41 |
|
[82] |
| 2. |
PE_PGRS18 |
|
[83] |
| 3. |
PE_PGRS5 |
|
[31][59] |
| 4. |
PE_PGRS11 |
-
TLR-2 agonist
-
Pro-inflammatory cytokines are stimulated
-
Dendritic cells are activated, which stimulate CD4+ T-cells
|
[50] |
| 5. |
PE_PGRS17 |
-
TLR-2 agonist
-
Pro-inflammatory cytokines are stimulated
-
Dendritic cells are activated, which stimulate CD4+ T-cells
|
[50] |
| 6. |
PE_PGRS33 |
|
[84] |
| 7. |
PE_PGRS62 |
|
[85] |
Table 4. Comprehensive table showing role of different PE/PPE paired proteins in host immune modulation.
| Sr. No. |
PE/PPE Proteins |
Role in Immune Modulation |
Reference |
| 1. |
PE32/PPE65 |
|
[86] |
| 2. |
PE9/PE10 |
|
[87] |
| 3. |
PE25/PPE41 |
|
[88] |
| 4. |
PE35/PPE68 |
|
[89] |