Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Tatiana Montoya Garcia | + 2892 word(s) | 2892 | 2021-12-28 10:32:07 | | | |
| 2 | Vivi Li | Meta information modification | 2892 | 2022-01-13 10:27:05 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Montoya Garcia, T.; Alarcòn De-La-Lastra, C.; Castejon, M.L.; Ortega-Vidal, J.; Altarejos, J.; Sánchez-Hidalgo, M. (−)-Methyl-Oleocanthal, a New Oleocanthal Metabolite. Encyclopedia. Available online: https://encyclopedia.pub/entry/18187 (accessed on 07 March 2026).
Montoya Garcia T, Alarcòn De-La-Lastra C, Castejon ML, Ortega-Vidal J, Altarejos J, Sánchez-Hidalgo M. (−)-Methyl-Oleocanthal, a New Oleocanthal Metabolite. Encyclopedia. Available at: https://encyclopedia.pub/entry/18187. Accessed March 07, 2026.
Montoya Garcia, Tatiana, Catalina Alarcòn De-La-Lastra, Maria Luisa Castejon, Juan Ortega-Vidal, Joaquin Altarejos, Marina Sánchez-Hidalgo. "(−)-Methyl-Oleocanthal, a New Oleocanthal Metabolite" Encyclopedia, https://encyclopedia.pub/entry/18187 (accessed March 07, 2026).
Montoya Garcia, T., Alarcòn De-La-Lastra, C., Castejon, M.L., Ortega-Vidal, J., Altarejos, J., & Sánchez-Hidalgo, M. (2022, January 13). (−)-Methyl-Oleocanthal, a New Oleocanthal Metabolite. In Encyclopedia. https://encyclopedia.pub/entry/18187
Montoya Garcia, Tatiana, et al. "(−)-Methyl-Oleocanthal, a New Oleocanthal Metabolite." Encyclopedia. Web. 13 January, 2022.
Copy Citation
The antioxidant and anti-inflammatory responses of (−)-methyl-oleocanthal (met-OLE), a new metabolite of the extra virgin olive oil (EVOO) phenolic oleocanthal (OLE), were explored in lipopolysaccharide (LPS)-induced murine peritoneal macrophages. Possible signaling pathways and epigenetic modulation of histones were studied. Met-OLE inhibited LPS-induced intracellular reactive oxygen species (ROS) and nitrite (NO) production and decreased the overexpression of the pro-inflammatory enzymes COX-2, mPGES-1 and iNOS in murine macrophages.
antioxidant
histones
inflammation
macrophages
metabolite
methylation
oleocanthal
olive oil
1. Introduction
Macrophages are major components of the innate immune system and play a critical role in modulating inflammatory and immune responses [1]. Extracellular bacterial lipopolysaccharide (LPS) acts as pathogen-associated molecular pattern and is recognized by the Toll-like receptor (TLR)-4, inducing macrophages to an activated state, producing pro-inflammatory cytokines and chemokines and enhancing the expression of inflammatory-related enzymes, such as inducible nitric oxide synthase (iNOS), cyclooxygenase (COX)-2 and microsomal prostaglandin E synthase (mPGES)-1, which synthesize nitric oxide (NO) and prostaglandin (PG)E2, respectively [2]. Additionally, LPS-stimulated macrophages disrupt the balance of the intracellular reduction–oxidation state, leading to oxidative stress, usually accompanied by damage that is mediated by reactive oxygen species (ROS) [3]. The process of gene expression of these pro-inflammatory mediators involves multiple signal transduction pathways, which are mainly through mitogen-activated protein kinases (MAPKs), nuclear transcription factor-kappa B (NF-κB), janus kinase/signal transducer and transcription activator of transcription (JAK/STAT) or inflammasome activation. Furthermore, the nuclear factor (erythroid-derived 2)-like 2 (Nrf-2)/haem oxygenase-1 (HO-1) antioxidative axis, which exerts a regulative function on the activation of ROS, MAPKs, and inflammasome signaling pathways, is repressed in the event of the induction of the activated macrophages state [2][3].
Emerging evidence suggests that epigenetic processes that affect gene expression without causing changes in the nucleotide sequence occur after external stimuli exposure, and may contribute to the pathophysiology of inflammatory processes [4]. In particular, histone H3 methylation at lysine 9 (H3K9), one of the most conserved epigenetic markers, is correlated with gene silencing and the modulation of immune cell differentiation and immune responses, and therefore, influences the outcome of inflammation. Similarly, H3 acetylation on lysine 18 (H3K18ac) is a permissive marker on genes encoding cytokines that correlate to inflammation, such as interleukin (IL)-1β, IL-6 or IL-17 [5][6][7]. Furthermore, post-translational histone modifications have emerged as prospective therapeutic targets. Understanding the cross-link of these mechanisms could be crucial in designing new immune–inflammatory approaches that are effectively related to several inflammatory diseases [8].
In this context, dietary nutrients could modify physiological and pathological processes through critical epigenetic mechanisms, promoting modifications of gene expression without alteration of the genetic code. In particular, specific, functional foods such as extra virgin olive oil (EVOO) have displayed anti-inflammatory activities in human macrophages through epigenetic mechanisms [9].
The health-promoting properties of EVOO have been correlated with its peculiar chemical composition. EVOO polyphenols are minor secondary metabolites that have been widely studied due to their wide functional versatility, including their anti-inflammatory, antioxidant, cardioprotective, chemopreventive, and neuroprotective properties [10]. Essentially, the anti-inflammatory activities of olive oil, especially phenolic compounds, have recently been linked to their potential to induce epigenetic modifications such as gene expression, DNA methylation and histone modification [10][11].
Examples of the main olive polyphenols are tyrosol, hydroxytyrosol, oleocanthal (OLE), oleacein, olive ligustroside and oleuropein. OLE represents up to 10% of the total polyphenol content in EVOO (0.2–498 mg/kg) [12] and has received more scientific attention, due to its interesting biological activities, both within in vitro and in vivo systems, including anti-inflammatory, antioxidant, cardioprotective, chemopreventive and neuroprotective properties [10][13]. In fact, recently, we have reported the preventive role of dietary OLE-supplemented effects in a collagen-induced arthritis (CIA) murine model and the ability of OLE to diminish the acute inflammatory response in LPS-induced murine peritoneal macrophages [14][15].
Metabolic transformations or the presence of metabolites can affect the pharmacological activity of the pattern compounds. It has been reported that OLE can remain intact in the stomach for up to 4 h and enter the small intestine unhydrolyzed. Then, non-hydrolyzed OLE follows further metabolic reactions related to phase I and II in the liver, namely, hydroxylation or hydration and methylation, respectively. Consequently, López-Yerena et al. (2021) proposed two plausible metabolites of OLE, however, met-OLE was the main circulating conjugate of OLE detected in all tissues analyzed from rats after the acute intake of a refined olive oil containing 0.3 mg/mL of OLE [16]. In relation to biotransformation, the methylated metabolites are mainly metabolized by glucuronidation and sulphation, as they are not substrates for methyltransferases.
Perhaps, the major disadvantage of the EVOO phenols is their temperature instability, photolability, and inadequate pharmacokinetic profile. To reduce these handicaps, methylation of the phenolic hydroxyl groups (O-methylation) may increase the chemical stability of their structures, while conferring greater lipophilicity, increasing their metabolic stability and membrane transport and facilitating absorption and greater oral bioavailability. Recent studies also indicate that the methylation process increases biological activity without altering therapeutic indices [17].
2. Chemistry
The synthesis of (−)-met-OLE was carried out herein from (−)-ligustroside (Figure 1), inspired by the reaction reported by Skaltsounis’ group for the conversion of oleuropein (same structure as ligustroside but with an additional OH at C-5′) into oleacein (same structure as OLE but with an additional OH at C-5′) [18]. These authors achieved the semi-synthesis of oleacein from oleuropein under Krapcho decarbomethoxylation conditions in one single step. They refluxed oleuropein, dissolved in wet DMSO, with two equivalents of an inorganic salt (NaCl) for 10 h to obtain oleacein with a 21% yield, after purification by silica gel column chromatography. Procopio’s group later improved this attractive semi-synthesis through changing DMSO by water and heating in a microwave reactor to get oleacein in just 20 min with a 48% yield [19]. Our group was also working to improve the semi-synthesis of oleacein and OLE from oleuropein and ligustroside, respectively, following the recommendations of a previous work aimed at adapting the classical Krapcho decarboxylation experimental conditions to an aqueous microwave scenario [20], when Procopio’s work came to light (unpublished results).
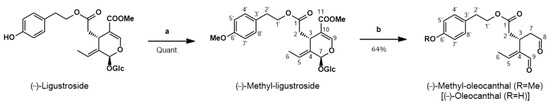
Figure 1. Synthesis of (−)-methyl-oleocanthal from (−)-ligustroside. Reagents and conditions: (a) CH2N2/Et2O; (b) H2O-DMSO, MW, 180 °C, 9 min.
Thus, our synthesis of met-OLE (Figure 1) started with the isolation of ligustroside from olive wood following a procedure previously reported by us [21]. Then, ligustroside was methylated using diazomethane, prepared in situ from N-methyl-N-nitrosourea. The conversion of ligustroside into methyl-ligustroside was quantitatively performed at room temperature for around 1 h. If the starting material (ligustroside) had been pure enough, methyl-ligustroside would have been sufficiently pure to be used directly in the next step. However, ligustroside was isolated with a purity of 84% from the olive wood extract and it was convenient to submit crude methyl-ligustroside to purification, in order to eliminate the minor components that accompanied the starting material (ligustroside). On this occasion, we used fast centrifugal partition chromatography (FCPC) to get adequately pure methyl-ligustroside in one single run. Then, pure methyl-ligustroside was submitted to the Krapcho reaction under microwave irradiation. Several attempts were carried out, with varying temperatures, reactions time and solvent ratios, until the best conditions were found. Thus, the reaction of methyl-ligustroside in water, with the minimum amount of DMSO to dissolve the substrate, was performed in a microwave reactor for 9 min at 180 °C, reaching the complete conversion of methyl-ligustroside into met-OLE. It is worth noting that this Krapcho reaction took place in the absence of inorganic salt, as Murphree’s group [20] came to observe in some cases and as Fernández-Bolaños’ group has recently claimed [22].
In order to purify the final compound and taking into account the known sensitivity of related OLE and oleacein to decomposition when in contact with solid stationary phases, such as silica gel [23], and upon exposure to oxygen and light [12], we used the FCPC technique for the purification of met-OLE. This allowed us to obtain pure met-OLE with the same success as when we previously purified OLE from an olive oil phenolic extract [24]. This silica-free chromatographic technique has shown to be a good option to isolate these compounds [25] and consequently improve the yield of the reaction step. Thus, we have performed the conversion of methyl-ligustroside into met-OLE in a 64% yield, which is the best yield reported to date for the Krapcho conversion of ligustroside/oleuropein or their derivatives into the corresponding dialdehydes. Met-OLE has been synthesized in this work for the first time.
3. Effects of Met-OLE on Cell Viability
First, we evaluated the cell viability after met-OLE treatments using a SRB assay. Data show that met-OLE was not cytotoxic after 18 h of treatment with concentrations of 1.6 up to 200 μM and did not compromise the cell viability significantly (≥80%) (Figure 2). Therefore, based on our previous work using OLE [14], we selected 12.5, 25 and 50 μM concentrations of met-OLE to be studied in the following assays.
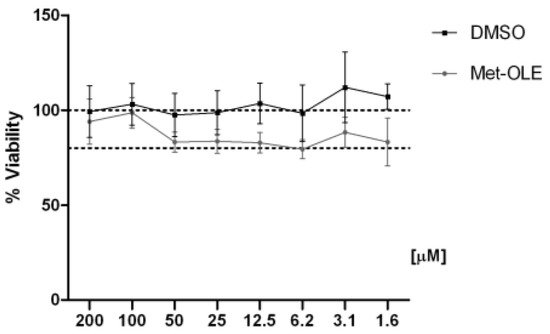
Figure 2. Effect of met-OLE on cell viability. Macrophages were pretreated with met-OLE (200–1.6 μM) for 18 h. Cell survivals were expressed as the percentage of viability with respect to 100% from control, untreated cells. Results are presented as mean ± SEM of at least six independent experiment.
4. Effects of Met-OLE on IL-1β, IL-6, IL-17, IFN-γ and TNF-α Production
To explore the effects of met-OLE on pro-inflammatory cytokine production, we evaluated IL-1β, IL-6, IL-17, IFN-γ and TNF-α levels. As shown in Figure 3, after 18 h of exposure to LPS, murine cells exhibited higher levels of pro-inflammatory cytokines than unstimulated control cells (++ p < 0.01; +++ p < 0.001 vs. unstimulated cells). On the contrary, when cells were treated with met-OLE, we observed a significantly down-regulation of IL-1β, IL-6, IL-17, IFN-γ and TNF-α secretions when compared to the LPS-DMSO group (** p < 0.01; *** p < 0.001 vs. LPS-DMSO stimulated cells).
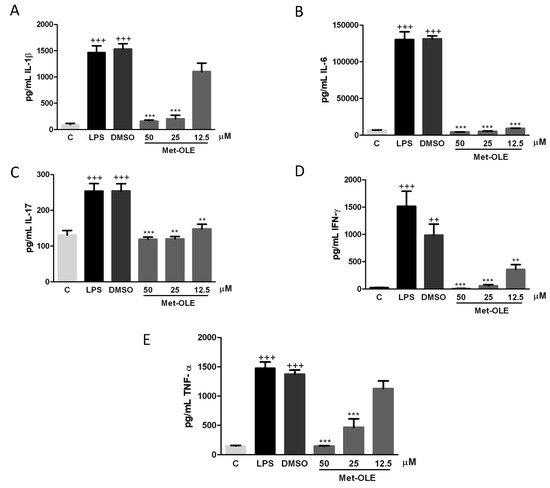
Figure 3. Pro-inflammatory cytokine levels were down-regulated in met-OLE treated cells. Macrophages were pretreated with met-OLE (50, 25 or 12.5 μM) for 30 min and then, LPS-stimulated during 18 h. (A) IL-1β, (B) IL-6, (C) IL-17, (D) IFN-γ and (E) TNF-α levels were measured by ELISA on cell supernatants. Results are presented as the mean ± SEM of at least six independent experiments. ++ p < 0.01; +++ p < 0.001 vs. unstimulated control cells; ** p < 0.01; *** p < 0.001 vs. LPS-DMSO stimulated cells.
5. Effects of Met-OLE on Intracellular ROS and NO Productions
In terms of elucidating the role of met-OLE in the oxidative and inflammatory response mediated by LPS, we measured intracellular ROS and NO levels using DCFDA and Griess assays, respectively, in LPS-induced murine peritoneal macrophages.
Data revealed remarkable overproductions of ROS and NO induced by LPS in murine macrophages when compared to unstimulated cells (+ p < 0.05; ++ p <0.01; +++ p < 0.001 vs. unstimulated control cells) (Figure 4A,B). Meanwhile, levels of both mediators were significantly reduced after met-OLE treatments (** p < 0.01; *** p < 0.001 vs. cells stimulated cells).
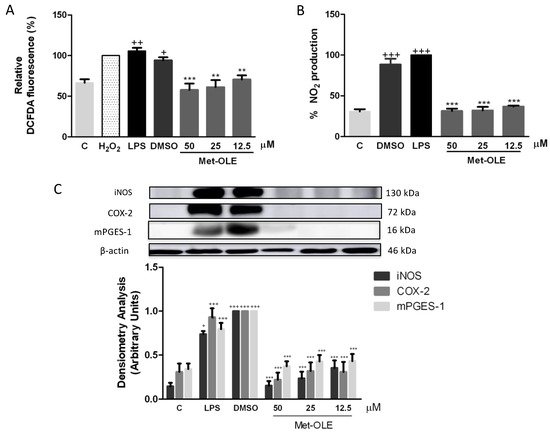
Figure 4. Effects of met-OLE on (A) intracellular ROS and (B) NO production and (C) overexpression of the iNOS, COX-2 and mPGES-1 proteins. Macrophages were pretreated with met-OLE (50, 25 or 12.5 μM) for 30 min and then, LPS-stimulated during 18 h. Subsequently, ROS and NO levels were measured using DCFDA and Griess assays in supernatants, respectively. The expression of iNOS, COX-2 and mPGES-1 proteins was then measured in whole cell lysates and analyzed by Western blotting. Results are presented as mean ± SEM of at least six independent experiments. + p < 0.05; ++ p < 0.01; +++ p < 0.001 vs. unstimulated control cells; ** p < 0.01; *** p < 0.001 vs. LPS-DMSO stimulated cells.
6. Met-OLE Down-Regulated iNOS, COX-2 and mPGES-1 Protein Overexpression Induced by LPS in Murine Macrophages
To investigate whether the effects of met-OLE on NO accumulation were associated with the expression of the iNOS protein, we performed an immunoblotting assay with cell lysates. Consistently, cells from secoiridoid-related phenolic groups showed decreased iNOS protein overexpression when compared to those of the LPS-DMSO exposed group (*** p < 0.001 vs. LPS-DMSO stimulated cells) (Figure 4C).
In addition, to gain further insight into the anti-inflammatory potential of met-OLE, we studied its effects on COX-2 and PGE2 related biomarkers. As expected, after LPS exposure, COX-2 and mPGES-1 expressions increased significantly (+++ p < 0.001 vs. unstimulated control cells). On the contrary, met-OLE pretreatments effectively counteracted induction of the expressions of these proinflammatory enzymes (*** p < 0.001 vs. LPS-DMSO stimulated cells) (Figure 4C).
7. Effects of Met-OLE on LPS-Induced MAPKs Activation in Murine Peritoneal Macrophages
Murine macrophages were previously pretreated with met-OLE and were LPS-induced. After 18 h, cells from the LPS-DMSO control group showed a marked phosphorylation of MAPK members: p38, ERK and JNK (+ p < 0.05; ++ p < 0.01; +++ p < 0.001 vs. unstimulated cells). Nevertheless, met-OLE counteracted the activation of these proteins and exhibited a lower phosphorylation for p38, ERK and JNK MAPK (* p < 0.05; ** p < 0.01; *** p < 0.001 vs. LPS-DMSO stimulated cells (Figure 5A).
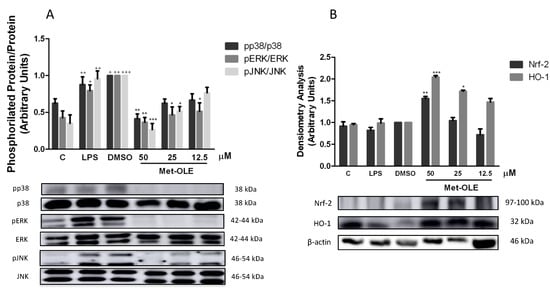
Figure 5. Met-OLE inhibited (A) MAPKs phosphorylation and (B) upregulated the Nrf-2/HO-1 antioxidation axis in LPS-induced cells. Macrophages were pretreated and stimulated with LPS during 18 h with met-OLE (50, 25 or 12.5 μM). Subsequently, MAPKs, Nrf-2, and HO-1 protein expressions were measured in whole cell lysates by Western blot. Results are presented as the mean ± SEM of at least six independent experiments. + p < 0.05; ++ p < 0.01; +++ p < 0.001 vs. unstimulated control cells; * p < 0.05; ** p < 0.01; *** p < 0.001 vs. LPS-DMSO stimulated cells.
8. Effects of Met-OLE on Nrf2-Mediated Transcriptional Activation and HO-1 Induction in LPS Murine Peritoneal Macrophages
To identify whether the OLE derivative evoked an antioxidant effect on cells, we studied the Nrf-2/HO-1 axis, measuring both protein expressions by immunoblotting. As shown in Figure 5B, Nrf-2 and HO-1 protein expression was significantly up-regulated in met-OLE-exposed cells when compared to LPS-DMSO controls (* p < 0.05; ** p < 0.01; *** p < 0.001 vs. LPS-DMSO stimulated cells). The expression of the antioxidant Nrf-2/HO-1 protein that was induced was according to the ROS reduction observed in Figure 4A. Altogether our data highlighted the high antioxidant potential of met-OLE.
9. Effects of Met-OLE on Canonical and Noncanonical Inflammasome Signaling Pathways
In the presence of LPS, the inflammasome induces the inflammatory response in murine peritoneal macrophages [14][26]. Unsurprisingly, after 18 h of LPS exposition, cells showed a marked induction of canonical and noncanonical inflammasome implicates (+ p < 0.05; ++ p < 0.01; +++ p < 0.001 vs. unstimulated cells). According to the canonical pathway, we observed that met-OLE modulated the protein levels of nucleotide-binding oligomerization domain-like receptor protein 3 (NLRP3) and apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC), which activated pro-caspase-1 and caspase-1. All of these inflammasome canonical pathway proteins were effectively negatively regulated in pretreated macrophages (Figure 6).
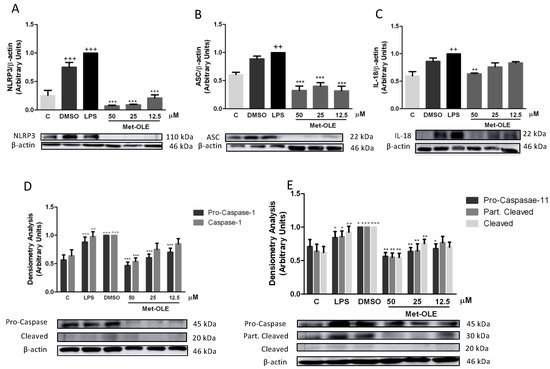
Figure 6. Met-OLE modulated the canonical and noncanonical inflammasome signaling pathways. Cells were pretreated with met-OLE (50, 25 or 12.5 μM) for 30 min and then, LPS-stimulated during 18 h. (A) NLRP3, (B) ASC, (C) IL-18, (D) Pro- and caspase-1 and (E) Pro-, partially cleaved and cleaved caspase-11 protein expressions were evaluated by Western blot. Results are presented as the mean ± SEM of at least six independent experiments. + p < 0.05; ++ p < 0.01; +++ p < 0.001 vs. unstimulated cells; * p < 0.05; ** p < 0.01; *** p < 0.001 vs. LPS-DMSO stimulated cells.
The protein expressions of the non-canonical pathway were then analyzed. As shown in Figure 5E, the expressions of pro-caspase-11, partially cleaved and cleaved caspase-11 were significantly reduced in cells incubated with met-OLE incubated cells (* p < 0.05; ** p < 0.01; *** p < 0.001 vs. LPS-DMSO stimulated cells). Lastly, in accordance with previous results, the expression levels of the mature form of IL-18 were measured, showing a significant reduction after met-OLE 50 μM treatment (** p < 0.01 vs. LPS-DMSO stimulated cells) (Figure 6C).
Collectively, met-OLE exhibited significant regulatory effects in LPS-induced canonical and non-canonical inflammasome signaling pathways in murine peritoneal macrophages.
10. OLE and Met-OLE Induced Epigenetic Histone Modifications
In an attempt to elucidate the role of epigenetic histone mechanisms underlying met-OLE’s anti-inflammatory effects, we explored met-OLE-induced epigenetic changes in histone markers and cytokine-correlated production, compared to OLE. We isolated histones from mice spleen cells and measured their expression by Western blot. A total of three H3 histone modifications were evaluated on several lysine residues: H3K18ac, H3K9me3 and H3K27me3. Acetylation of H3 is associated with the induction of expression, while methylation is a repressive marker. Consistent with that, basal levels of H3K18ac were induced, as well as H3K9me3 and H3K27me3 being significantly decreased after exposure to LPS exposition (+ p < 0.05; ++ p < 0.01 vs. unstimulated cells). Surprisingly, as shown in Figure 7A, pretreated cells with OLE and met-OLE counteracted LPS induction, preventing H3K18 acetylation or H3K9 and H3K27 demethylation (* p < 0.05; ** p < 0.01 vs. LPS-DMSO stimulated cells). Subsequently, we next evaluated the levels of several modulated cytokines that have been associated with the markers of these histones in their promoter regions. Particularly, levels of IL-17, IL-1β and IL-6 were up-regulated after LPS stimulation (+ p < 0.05; ++ p < 0.01; +++ p < 0.001 vs. unstimulated cells), while the treatments with OLE and met-OLE were able to considerably reduce them. These results with previous results obtained for the histones’ modifications (* p < 0.05; ** p < 0.01; *** p < 0.001 vs. LPS-DMSO stimulated cells) (Figure 7B).
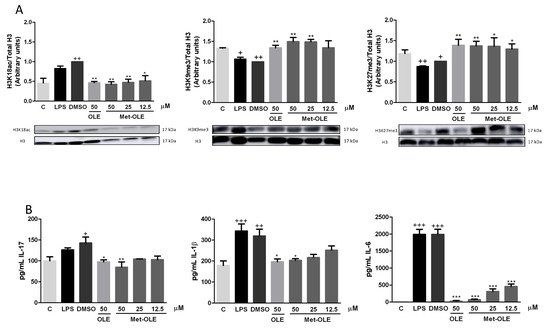
Figure 7. (A) Epigenetic modifications of histones by OLE and met-OLE and (B) the cytokine-related response. Spleen cells were pre-treated with OLE (50 μM) or met-OLE (50, 25 or 12.5 μM) for 30 min and then, LPS-stimulated during an 18 h period. The histones were isolated from cells and the expression of H3 modifications was evaluated using antibodies for H3K18ac, H3K9me3 and H3K27me3 by Western blot. Related cytokine levels were measured in cell supernatants using specific ELISA assays. Results are presented as the mean ± SEM of at least six independent experiments. + p < 0.05; ++ p < 0.01; +++ p < 0.001 vs. unstimulated cells; * p < 0.05; ** p < 0.01; *** p < 0.001 vs. LPS-DMSO stimulated cells.
Interestingly, all together the results suggest that OLE and met-OLE exert effects on epigenetic regulation in acute inflammatory responses and innate immunity, reducing H3K18 acetylation and inducing H3K9 and H3K27 methylation on il17, il1β and il6 promoters, and probably suppressing their transcription.
References
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455.
- Yao, Y.; Liu, K.; Zhao, Y.; Hu, X.; Wang, M. Pterostilbene and 4′-Methoxyresveratrol Inhibited Lipopolysaccharide-Induced Inflammatory Response in RAW264.7 Macrophages. Molecules 2018, 23, 1148.
- Brüne, B.; Dehne, N.; Grossmann, N.; Jung, M.; Namgaladze, D.; Schmid, T.; Von Knethen, A.; Weigert, A. Redox Control of Inflammation in Macrophages. Antioxid. Redox Signal. 2013, 19, 595–637.
- Shanmugam, M.K.; Sethi, G. Role of Epigenetics in Inflammation-Associated Diseases. Epigenetics Dev. Dis. 2013, 61, 627–657.
- Hajji, N.; Joseph, B. Epigenetic regulation of cell life and death decisions and deregulation in cancer. Essays Biochem. 2010, 48, 121–146.
- Zhao, S.; Zhong, Y.; Fu, X.; Wang, Y.; Ye, P.; Cai, J.; Liu, Y.; Sun, J.; Mei, Z.; Jiang, Y.; et al. H3K4 Methylation Regulates LPS-Induced Proinflammatory Cytokine Expression and Release in Macrophages. Shock 2019, 51, 401–406.
- Imuta, H.; Fujita, D.; Oba, S.; Kiyosue, A.; Nishimatsu, H.; Yudo, K.; Suzuki, E. Histone methylation and demethylation are implicated in the transient and sustained activation of the interleukin-1β gene in murine macrophages. Hear. Vessel. 2020, 35, 1746–1754.
- Hachiya, R.; Shiihashi, T.; Shirakawa, I.; Iwasaki, Y. The H3K9 methyltransferase Setdb1 regulates TLR4-mediated inflam-matory responses in macrophages. Sci. Rep. 2016, 28, 28845.
- Bordoni, L.; Fedeli, D.; Fiorini, D.; Gabbianelli, R. Extra Virgin Olive Oil and Nigella sativa Oil Produced in Central Italy: A Comparison of the Nutrigenomic Effects of Two Mediterranean Oils in a Low-Grade Inflammation Model. Antioxidants 2019, 9, 20.
- Castejón, M.L.; Montoya, T.; Alarcón-De-La-Lastra, C.; Sánchez-Hidalgo, M. Potential Protective Role Exerted by Secoiridoids from Olea europaea L. in Cancer, Cardiovascular, Neurodegenerative, Aging-Related, and Immunoinflammatory Diseases. Antioxidants 2020, 9, 149.
- Fabiani, R.; Vella, N.; Rosignoli, P. Epigenetic Modifications Induced by Olive Oil and Its Phenolic Compounds: A Systematic Review. Molecules 2021, 26, 273.
- Cicerale, S.; Conlan, X.; Barnett, N.W.; Keast, R. Storage of extra virgin olive oil and its effect on the biological activity and concentration of oleocanthal. Food Res. Int. 2013, 50, 597–602.
- Segura-Carretero, A.; Curiel, J.A. Current Disease-Targets for Oleocanthal as Promising Natural Therapeutic Agent. Int. J. Mol. Sci. 2018, 19, 2899.
- Montoya, T.; Castejón, M.L.; Sánchez-Hidalgo, M.; González-Benjumea, A.; Fernández-Bolaños, J.G.; De-La-Lastra, C.A. Oleocanthal Modulates LPS-Induced Murine Peritoneal Macrophages Activation via Regulation of Inflammasome, Nrf-2/HO-1, and MAPKs Signaling Pathways. J. Agric. Food Chem. 2019, 67, 5552–5559.
- Montoya, T.; Sánchez-Hidalgo, M.; Castejón, M.; Rosillo, M.; González-Benjumea, A.; Alarcón-De-La-Lastra, C. Dietary Oleocanthal Supplementation Prevents Inflammation and Oxidative Stress in Collagen-Induced Arthritis in Mice. Antioxidants 2021, 10, 650.
- López-Yerena, A.; Vallverdú-Queralt, A.; Jáuregui, O.; Garcia-Sala, X.; Lamuela-Raventós, R.; Escribano-Ferrer, E. Tissue Distribution of Oleocanthal and Its Metabolites after Oral Ingestion in Rats. Antioxidants 2021, 10, 688.
- Wen, L.; Jiang, Y.; Yang, J.; Zhao, Y.; Tian, M.; Yang, B. Structure, bioactivity, and synthesis of methylated flavonoids. Ann. N. Y. Acad. Sci. 2017, 1398, 120–129.
- Vougogiannopoulou, K.; Lemus, C.; Halabalaki, M.; Pergola, C.; Werz, O.; Smith, A.B., III; Michel, S.; Skaltsounis, L.; Deguin, B. One-step semisynthesis of olea-cein and the determination as a 5-lipoxygenase inhibitor. J. Nat. Prod. 2014, 77, 441–445.
- Costanzo, P.; Bonacci, S.; Cariati, L.; Nardi, M.; Oliverio, M.; Procopio, A. Simple and efficient sustainable semi-synthesis of oleacein as potential additive for edible oils. Food Chem. 2018, 245, 410–414.
- Mason, J.D.; Murphree, S.S. Microwave-Assisted Aqueous Krapcho Decarboxylation. Synlett 2013, 24, 1391–1394.
- Pérez-Bonilla, M.; Salido, S.; van Beek, T.A.; Linares-Palomino, P.J.; Altarejos, J.; Nogueras, M.; Sánchez, A. Isolation and identification of radical scavengers in olive tree (Olea europaea) wood. J. Chromatogr. A 2006, 1112, 311–318.
- Fernández-Bolaños, J.M.; Maya-Castilla, I.; González-Benjumea, A. Use of DMSO for the Synthesis of Oleacein and Oleocanthal. WO2018162769A2, 13 September 2018.
- Karkoula, E.; Skantzari, A.; Melliou, E.; Magiatis, P. Quantitative Measurement of Major Secoiridoid Derivatives in Olive Oil Using qNMR. Proof of the Artificial Formation of Aldehydic Oleuropein and Ligstroside Aglycon Isomers. J. Agric. Food Chem. 2014, 62, 600–607.
- Diez-Bello, R.; Jardin, I.; Lopez, J.; El Haouari, M.; Ortega-Vidal, J.; Altarejos, J.; Salido, G.; Salido, S.; Rosado, J. (−)-Oleocanthal inhibits proliferation and migration by modulating Ca2+ entry through TRPC6 in breast cancer cells. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2019, 1866, 474–485.
- Angelis, A.; Michailidis, D.; Antoniadi, L.; Stathopoulos, P.; Tsantila, V.; Nuzillard, J.-M.; Renault, J.-H.; Skaltsounis, L.A. Pilot continuous centrifugal liquid-liquid extraction of extra virgin olive oil biophenols and gram-scale recovery of pure oleocanthal, oleacein, MFOA, MFLA and hydroxytyrosol. Sep. Purif. Technol. 2021, 255, 117692.
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.M.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. JNCI J. Natl. Cancer Inst. 1990, 82, 1107–1112.
More
Information
Subjects:
Chemistry, Organic; Nutrition & Dietetics
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
2 times
(View History)
Update Date:
13 Jan 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





