Most defensins are cationic antimicrobial peptides with broad-spectrum killing activity against bacteria, fungi and enveloped viruses. However, it should be recognized that there are some non-cationic β-defensins in organisms, which need to be further studied. In this study, a new spliced isoform of anionic β-defensin from flounder (Paralichthys olivaceus, fBD) was identified, and its antibiosis, chemotaxis and modulation of phagocytosis were examined. In addition, the contributions of fBD to the antimicrobial activity of extracellular traps (ETs) were also analyzed. The recombinant fBD (rfBD) could effectively inhibit the growth of Gram-positive bacteria (S. aureus, Micrococcus luteus) and Gram-negative bacteria (E. coli, V. alginolyticus, V. anguillarum).
1. Introduction
The immune system of vertebrates is divided into two parts: innate immunity and adaptive immunity. As the first line of defense of the immune system, innate immunity fulfills powerful functions and plays an important role. For lower vertebrates such as bony fish, the innate immune system is particularly important because the formation of an adaptive immune response takes a long time and is affected by the ambient temperature
[1]. Antimicrobial peptides, are small-molecule peptides, also known as host defense peptides, usually consisting of 10–60 amino acids, with the characteristics of high temperature resistance, strong cationicity and hydrophobicity
[2].
Bony fish, as aquatic temperature-changing vertebrates, are directly exposed to the water environment rich in microorganisms. When the body is damaged or invaded by pathogenic microorganisms, the innate immune system is quickly activated, releasing AMPs, which are important substances to perform its functions of antibiosis. Up to now, fish antimicrobial peptides have been primarily divided into piscidins, defensins, hepcidins, cathelicidins and NK-lysins
[3]. Defensins are short, cysteine-rich cationic host defense peptides, divided into α-, β- and θ-defensins in vertebrates
[4]. The disulfide bond pairing methods and the positions of the six cysteine residues forming the disulfide bonds of defensins are different
[5]. The three disulfide bonds of α-defensin are connected by C1–C6, C2–C4 and C3–C5, while the pairing method of the six cysteines of β-defensin is C1–C5, C2–C4 and C3–C6. In addition, θ-defensin, which also contains three pairs of disulfide bonds, is a cyclic peptide that has only been identified in non-human primates, such as rhesus monkeys
[6]. In vertebrates with a low degree of evolution (such as birds and fish), merely β-defensins have been identified, and none of the α-defensins or θ-defensins has been discovered
[7]. In fish, zebrafish β-defensin was first identified, and multiple defensin-like genes were subsequently reported in many other species
[8][9][10][11][12][13][14].
Most β-defensins are cationic antimicrobial peptides with broad-spectrum killing activity against bacteria, fungi and enveloped viruses
[15]. In addition to these antimicrobial activities, β-defensin participates in the interaction between the organism and microorganisms by regulating the phagocytic activity of phagocytes and the chemotaxis of immune cells. As a bridge between adaptive and innate immunity, β-defensin is a potential immune booster candidate for vaccines
[16][17]. In teleosts, some studies have demonstrated that cationic β-defensins contributed to the antibiosis
[18], inflammatory inhibition
[19], leukocytic chemotaxis
[20], modulation of phagocytosis and boost of viral DNA vaccines
[21].
In higher vertebrates, after being stimulated, several types of immune cells release chromatin and granular proteins into the extracellular space, forming DNA traps
[22]. This process occurs in many innate immune cells and is especially prominent in neutrophils. Its components including DNA, histones, elastase, myeloperoxidase, lactoferrin and defensins are involved in the regulation of innate and adaptive immunity
[23][24]. However, there was no report about the function and mechanism of defensin in neutrophils or extracellular traps in teleosts.
Flounder (
Paralichthys olivaceus) is an financially profitable teleost species extensively cultured in East Asian countries
[25]. Multiple anionic β-defensin isoforms from flounder have been identified, and the mature peptide’s antimicrobial activities have been found
[26]. Nevertheless, as an anionic antimicrobial peptide, it is poorly understood whether the function of β-defensin in flounder resembles that of cationic antimicrobial peptides. In this study, a new anionic β-defensin isoform from flounder (fBD) was identified, and its antibiosis, chemotaxis and modulation of phagocytosis were examined. In addition, the contributions of fBD to the antimicrobial function of extracellular traps were also analyzed.
2. Cloning and Sequence Analysis of fBD
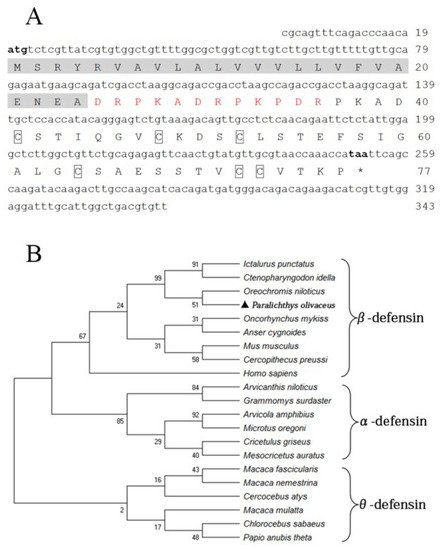
The fBD (GenBank accession No. OL631146) contained a 234 bp open reading frame (ORF) encoding 77 amino acids with a molecular weight of 13.25 kDa and a theoretical isoelectric point (pI) of 4.8. Six conserved cysteines were present in the amino acid sequence. The protein comprises a signal peptide (24 amino acids) with a pI of 4.0 and a mature peptide (53 amino acids) with a pI of 5.7, in theory (Figure 1A). The phylogenetic relationship of fBD to other species’ β-defensin genes was examined through an analysis of the ORF sequence. The fBD genes clustered with those of the Nile tilapia at a branch and clustered with those of channel catfish and grass carp at a high level of statistical confidence (Figure 1B).
Figure 1. Sequence structure and phylogenetic analysis of fBD. (A) Nucleotide and amino acid sequences of fBD. The initiation and termination codons are marked with bold letters. The signal peptide is marked with a highlight, the propiece region is shown in red, and fixed cysteines are framed by boxes. (B) Phylogenetic tree analysis of rfBD from flounder and other species. The tree was structured according to the “maximum-likelihood” method using MEGAX. The GenBank accession numbers of α-, β- and θ-defensins are listed as follows: Homo sapiens: AF529417.1; Ctenopharyngodon idella: MG652749.1; Oncorhynchus mykiss: NM_001195183.2; Anser cygnoides: EU606039.1; Mus musculus: AY591385.1; Oreochromis niloticus: MH674361.1; Ictalurus punctatus: KX211992.1; Cercopithecus preussi: EU126877.1; Arvicanthis niloticus: XM_034520497.1; Arvicola amphibius: XM_038325539.1; Cricetulus griseus: XM_027435861.1; Grammomys surdaster: XM_028784038.1; Mesocricetus auratus: XM_040733512.1; Microtus oregoni: XM_041673564.1; Cercocebus atys: XM_012078007.1; Chlorocebus sabaeus: XM_037984733.1; Macaca fascicularis: XM_005562541.2; Macaca mulatta: AF191100.1; Macaca nemestrina: XM_011711827.2; Papio anubis: FJ030939.1; Paralichthys olivaceus: OL631146.
3. rfBD Production and Antibody Verification
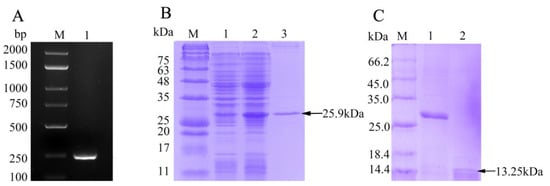
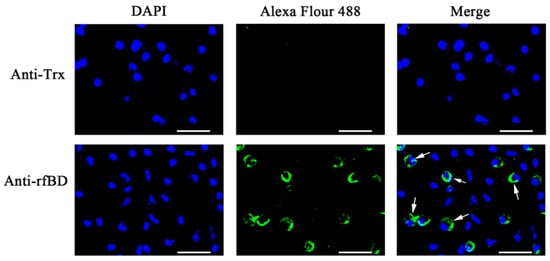
The ORF of fBD gene was cloned via PCR from the head kidney transcriptome (Figure 2A). The PCR product was successfully inserted into the vector pET-32a and verified using gene sequencing. rfBD fused with Trx tag was successfully induced and purified. The purified rfBD–Trx fusion protein was verified by SDS-PAGE (Figure 2B, theoretical size approximately 25.9 kDa). The Trx tag of the rfBD was successfully cleaved, and the rfBD without Trx tag was purified (Figure 2C, 13.25 kDa). Peritoneal cells of flounder were used to verify the specificity of the antibodies via immunofluorescence microscopy. As shown in Figure 3, the prepared antibody can be perfectly combined with the natural fBD protein in the cytoplasm.
Figure 2. The preparation of recombinant fBD. (
A) The agarose gel electrophoresis of fBD gene amplified from head kidney by PCR. Lane M, DNA marker; lane 1, PCR products of fBD. (
B) SDS-PAGE analysis of rfBD peptide. Lane M, protein marker; lane 1, uninduced whole-bacteria lysate; lane 2, induced whole-bacteria lysate; lane 3, purification of rfBD peptide with Trx tag. (
C) SDS-PAGE analysis of rfBD peptide after digestion. Lane M, protein marker; lane 1, purification of rfBD peptide with Trx tag; lane 2, purification of rfBD peptide without Trx tag. Full figure of PAGEs can be found in
Supplementary Materials (could be found in https://www.mdpi.com/2079-7737/10/12/1247#supplementary).
Figure 3. The localization of fBD in peritoneal cells. After incubation with anti-rfBD or anti-Trx antibody, the peritoneal cells were detected using Alexa Flour 488–labeled secondary antibody. The nucleus was visualized using DAPI. In both cases, the right panels were merges of the left and middle panels. The arrows indicate some representative fBD-positive cells. Bar, 20 μm.
4. Antimicrobial Activity of rfBD
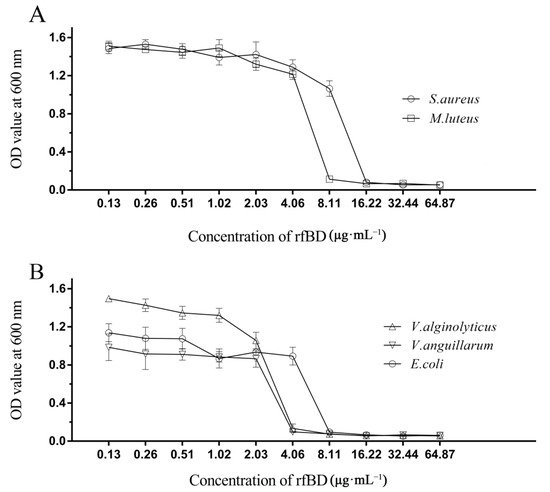
The dilution method was used to establish the MICs of rfBD against bacteria. rfBD showed significant concentration-dependent growth-inhibitory activity against S. aureus, M. luteus, E. coli, V. alginolyticus and V. anguillarum (Figure 4, Table 2). The MIC values are summarized in Table 2.
Figure 4. Antibacterial activities of rfBD. The broth dilution method was used to study the inhibitory effects of different concentrations of rfBD on two Gram-positive bacteria (
A) and three Gram-negative bacteria (
B). The results were expressed as the absorbance value of the culture solution at 600 nm (The data was summarized in
Table S1). The values in the graph are expressed as mean ± SD (
n = 3).
Table 2. Antimicrobial activities of rfBD determined by liquid growth inhibition assay. * The MIC values were noted as a scope among the highest concentration of the protein at which bacterial growth was observed and the lowest concentration that resulted in 100% inhibition of bacterial growth.
| Microorganisms |
MIC Value * (μg·mL−1) |
| Gram-positive bacteria |
|
| S. aureus |
8.11–16.22 |
| M. luteus |
4.06–8.11 |
| Gram-negative bacteria |
|
| E. coli |
4.06–8.11 |
| V. alginolyticus |
2.03–4.06 |
| V. anguillarum |
2.03–4.06 |
5. Localization and Effect of fBD on the ETs
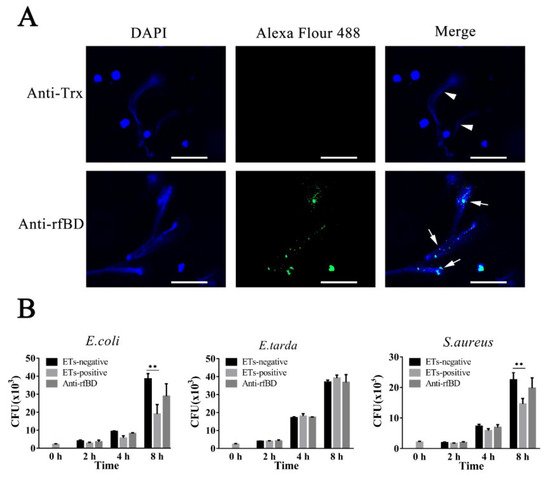
To examine whether the fBD was present in ETs, leukocytes of flounder were treated with the ET inducer PMA, and the resulting ETs were verified by immunofluorescence microscopy. As illustrated in Figure 5A, fBDs were detected by antibodies against rfBD co-localized with extracellular DNA fiber. Following the above results, we evaluated the functions of fBDs in conferring the antimicrobial activity of ETs. For this purpose, ET-positive cells were treated with or without antibodies against rfBD before being subjected to E. coli, Ed. tarda and S. aureus infection, and bacterial survival on ETs was monitored at 2, 4 and 8 h after infection. In E. coli and S. aureus infection groups, the recovery numbers of bacteria from ET-positive cells groups were significantly lower at 4 and 8 h compared to the ET-negative cells. The recovery rate of bacteria from ET-positive cells in the presence of antibodies against rfBD was increased compared to the ET-positive cells that were not treated, at 4 and 8 h after infection; however, there was no significant difference. In comparison, the number of Ed. tarda, collected from ET-positive cells with or without anti-rfBD antibodies were similar to those from ET-negative cells at all time points (Figure 5B).
Figure 5. The localization and antibacterial activities of fBD in extracellular traps. (A) PMA-stimulated ET-producing cells were treated with antibodies against Trx or rfBD followed by Alexa Fluor–labeled secondary antibody. Arrows without tail indicate ETs and arrows with tails indicate fBD in ETs. Bar, 20 μm. (B) Effect of rfBD on bacteriostatic capabilities of ETs. ET-producing cells were infected by E. coli, Ed. tarda and S. aureus with or without (control) of anti-rfBD, and bacterial survival was counted at different times. Data are the means of three independent assays and are presented as means ± SEM. ** p < 0.01.
6. Effects of rfBD on Chemotaxis of Leukocytes
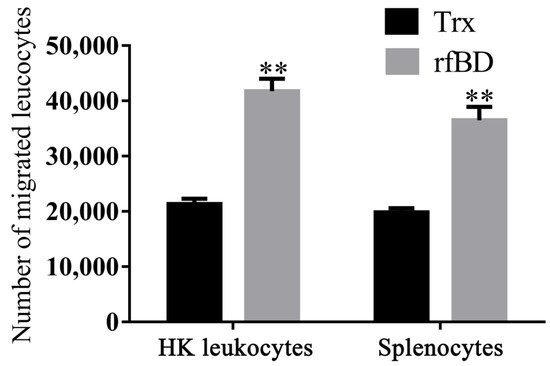
By using a transwell, rfBD was proved to have chemotactic ability to leucocytes from the head kidney and spleen. Flow cytometric results revealed that the number of leukocyte chemotaxes to the lower chamber was significantly increased in the presence of rfBD (Figure 6). The chemotactic number of leucocytes from the head kidney was 41,790 ± 2221 cells in the presence of rfBD, while in the control group, it was only 21,350 ± 968 cells. The chemotactic number of leucocytes from the spleen was 36,510 ± 2421 cells in the presence of rfBD, while in the control group, it was only 19,870 ± 728 cells.
Figure 6. Chemotactic effects of rfBD on leukocytes from head kidney and spleen in vitro. Migrated leukocytes to the lower chamber of the transwell were counted by flow cytometry. Bars represent the mean number of migrated leukocytes ± SEM (n = 3). The level of significance compared with the Trx treatment group is represented by ** p < 0.01.
7. Effects of rfBD on the Phagocytosis of Lymphocytes
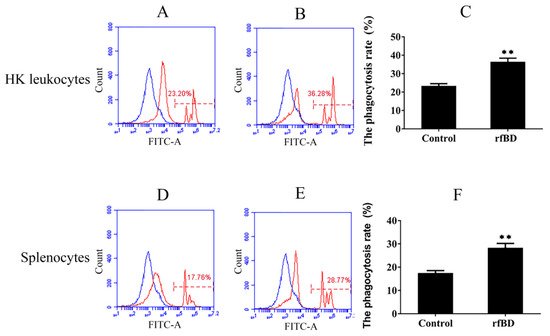
Phagocytosis assay was conducted to investigate whether rfBD can promote the phagocytic capability of leucocytes from the head kidney and spleen. Flow cytometric analysis revealed that leucocytes acquired significantly higher green-fluorescent-microsphere phagocytic activity after rfBD treatment (Figure 7). The phagocytic rate of leucocytes from the head kidney was 36.43% ± 2.02% in the group with rfBD treatment, while in the control group, it was only 23.31% ± 1.29%. The phagocytic rate of leucocytes from the spleen was 28.31% ± 1.90% in the group with rfBD treatment, while that in the control group was only 17.47% ± 1.09%.
Figure 7. The phagocytosis of leukocytes from head kidney and spleen with or without rfBD stimulation detected by flow cytometric analysis. FITC fluorescence histograms showed the ratio of leukocyte with (B,E) or without (A,D) rfBD stimulation. The phagocytosis rate of leukocytes from the head kidney (C) and the spleen (F), respectively, with rfBD stimulation was analyzed using SPSS. All the error bars represent the standard deviation of three biological replicates. ** p < 0.01.