Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Cenk Suphioglu | + 1829 word(s) | 1829 | 2021-12-30 03:02:55 | | | |
| 2 | Jessie Wu | Meta information modification | 1829 | 2022-01-11 06:33:55 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Suphioglu, C. Inhibition of the IL-4 Cytokine Pathway for Asthma. Encyclopedia. Available online: https://encyclopedia.pub/entry/18016 (accessed on 08 February 2026).
Suphioglu C. Inhibition of the IL-4 Cytokine Pathway for Asthma. Encyclopedia. Available at: https://encyclopedia.pub/entry/18016. Accessed February 08, 2026.
Suphioglu, Cenk. "Inhibition of the IL-4 Cytokine Pathway for Asthma" Encyclopedia, https://encyclopedia.pub/entry/18016 (accessed February 08, 2026).
Suphioglu, C. (2022, January 11). Inhibition of the IL-4 Cytokine Pathway for Asthma. In Encyclopedia. https://encyclopedia.pub/entry/18016
Suphioglu, Cenk. "Inhibition of the IL-4 Cytokine Pathway for Asthma." Encyclopedia. Web. 11 January, 2022.
Copy Citation
Allergies are a worldwide issue, with approximately 20% of the global population suffering from allergy-caused symptoms including rashes (atopic dermatitis), runny nose (allergic rhinitis), and life-threatening breathing problems (allergy-induced asthma).
interleukin 4 (IL-4)
interleukin 13 (IL-13)
1. Background
Asthma is commonly defined as smooth muscle contractions and chronic inflammation of the trachea and other associated airways, and can be caused by a variety of pathways involving gene and environmental interactions [1]. Research has revealed that over 100 genes are involved with pathways leading to asthma, but no single gene has been identified as the sole culprit, making treating asthma at its cause an arduous process [2]. Due to this, treatments for asthma have been heavily focused on the relief of symptoms on a day-to-day schedule, rather than addressing the underlying causes. These treatments are typically inhaled corticosteroids for short-term relief, or longer-lasting therapeutics such as long-acting β2–adrenergic receptors, leukotriene receptor antagonists, long-acting muscarinic antagonists, and for the most severe of symptoms, IgE-specific monoclonal antibody immunotherapy, also called omalizumab [3]. These treatments are all directed towards the relief of symptoms, rather than addressing the underlying pathways dysregulated in asthma. Of the standard therapeutic treatments, some are more successful in certain individuals than others. Certain trials have concluded that for 20% of their patients, asthma remained uncontrolled when treated with traditional corticosteroids [4].
The interleukin 4 and 13 pathways play integral roles in both type 1 allergy hypersensitivity and type 2 immunity [5][6]. Due to the integral role that the IL-4 pathway plays in allergen-induced asthma, it is a prime candidate for inhibitory drugs to treat allergic asthma.
2. IL-4 and IL-13
Interleukins 4 and 13 are key cytokines in allergic inflammation, and are secreted by numerous immune cells, most commonly T-helper-2 lymphocytes [7]. The release of these cytokines causes the production of immunoglobulin E [IgE] by B lymphocytes, as well as causing a range of effects in other cells. The introduction of IgE to mast cells causes an immediate allergic inflammation and response. IL-4 plays another role in asthma through the induction of mucin gene expression, causing the hypersecretion of mucus from goblet cells within the airways. The IL-4 cytokine induces all these processes and more when released, causing the symptoms observed within allergic asthmatics.
When T-lymphocytes release IL-4, all cells carrying the receptor respond. While nearly all cells carry this receptor, overall cell responses differ between cell types due to the unique STAT6 transcription factor involved with this signaling pathway, which activates different intracellular transcription pathways dependent on that cell type. This allows for IL-4 to elicit a range of responses from a wide variety of cell types, making it the ideal candidate for therapeutic intervention. IL-13 differs from IL-4, as it is an effector cytokine that regulates smooth muscle contraction and mucus production. Similar to the IL-4 signaling system, IL-13 works extracellularly through cell membrane receptors.
The IL-4 signaling pathway involves numerous transmembrane peptides and intracellular signaling molecules, which produce unique responses dependent on cell type. The transmembrane receptor for IL-4 is IL-4Rα. IL-4Rα is expressed in most cells, although in low numbers. The role of this transmembrane peptide chain is to form a complex with IL-4, at which point it will recruit a secondary receptor chain. There are two secondary receptor chains, IL-2Ryc and IL-13Ra1, which are expressed within different cell types. Nonhematopoietic cells express IL-13Ra1, whereas IL-2Ryc is expressed in low levels or absent. The opposite of this is true for lymphocytes, which express IL-2Ryc, but little to no IL-13Ra1. The receptor complex formed between IL-4Rα and IL-2yc is herein referred to as the type 1 IL-4 complex, whereas the complex formed by IL-4Rα and IL-13Rα1 is herein referred to as the type 22 IL-4 receptor complex. Once the three signaling molecules have been recruited, the receptor has formed a functional complex, which then undergoes a conformational change, allowing for the activation of intracellular signaling machinery [8][9].
Once the receptor complex is formed and has undergone the conformational change to be functional, associated intracellular signaling molecules will undergo auto- and cross-phosphorylation, resulting in their activation. The intracellular machinery activated is largely dependent on the third signaling chain recruited to the transmembrane complex, although they all belong to a signaling family called the Jak kinases. The activation of these Jak kinases causes the phosphorylation of other kinases and critical Y residues within the intracellular domains of IL-4Rα. Once phosphorylation has occurred, these Y residues function as “docking sites” for intracellular signaling molecules, most commonly STAT6. STAT6 is an intracellular signaling molecule that grants the IL-4 pathway the unique property of producing different cellular responses dependent on cell type. Upon contact with the activated IL-4 signaling complex, STAT6 molecules homodimerize and translocate to the nucleus, binding to specific DNA sequences to influence the cells’ overall expression. The IL-4 signaling pathway is not only involved with transcriptional change, but is also involved with the downregulation of elicited signals. Pathways that downregulate signaling pathways are increased by IL-4 activation, limiting the duration of the IL-4 and associated signals.
The IL-13 pathway shares many functional similarities with the IL-4 pathway. IL-13 has two receptors similar to the IL-4 pathway, but differs by utilizing two separate binding chains. The type of receptor that is formed is dependent on which chain the IL-13 molecule binds to, unlike IL-4, which is determined after binding. The IL-13 pathway also affects the IL-4 pathway, as studies have displayed the complex formed by IL-13 and its type 1 alpha receptor (IL-13Rα1) lessen the intracellular effect of the type 2 IL-4 receptor complex, therefore reducing the cellular response. The IL-13 pathway concludes in the activation of STAT6 molecules and insulin receptor substrates (IRSs), which produce cellular responses at a transcription level. Unlike the IL-4 pathway, the IL-13 pathway is a poor activator for IRS molecules. Whilst STAT6 activates allergy symptoms, IRSs are involved with cellular proliferation. Both the IL-4 and IL-13 pathways play roles in the inflammation associated with allergies; however, they also play fundamental roles in cellular function [10][11][12] (Figure 1).
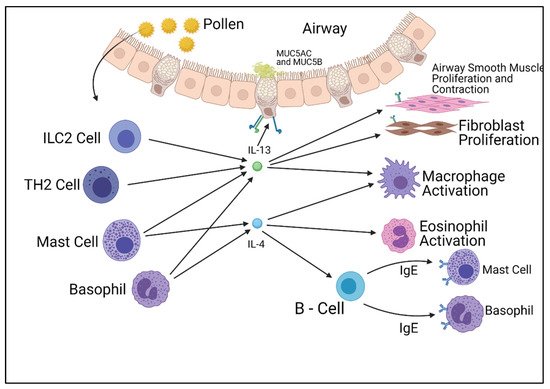
Figure 1. Summary of the cells responsible for the production of IL-4 and IL-13 and their effects on other cell types. Figure was created by the authors and reproduced with permission from BioRender.com (accessed on 19 November 2021).
STAT6 is activated by both the IL-4 and IL-13 pathways, though this is largely dependent on the cell type, as are the effects of the STAT6 molecule. IL-4, however, can induce further intracellular signaling pathways, including Sos/Ras, PI3K/Akt, PKB/mTOR, and PKC. These alternative pathways, coupled with the transcriptional influence of STAT6, create variable cellular responses to the introduction of the IL-4 or IL-13 cytokines. While having effects on many different cell types, the main functions of the IL-4 and IL-13 pathways are the production of human immunoglobulin E and T-helper cell differentiation, which then brings on the clinical symptoms of atopic dermatitis, allergic rhinitis, and allergic asthma.
STAT6 is a complex transcription factor responsible for exerting the effects of IL-4 and IL-13. Following phosphorylation by IL-4 and IL-13 receptor kinases, the phosphorylated STAT6 protein migrates to the nucleus to act as a transcription activator. It has been proven through mice knockout studies that STAT6 is required for allergy development; however, the genetic profile activated varies greatly between cell types. When STAT6 is activated within B-lymphocyte cells, STAT6 upregulates the Igε heavy chain and CD23 gene, which encodes for the IgE heavy chain and IgE receptors [13][14][15]. However, in Th2 cells, STAT6 upregulates gata3, another transcription factor responsible for many biological functions, and CRTH2, a gene involved in eosinophilic and allergic inflammation [16] (Figure 2).
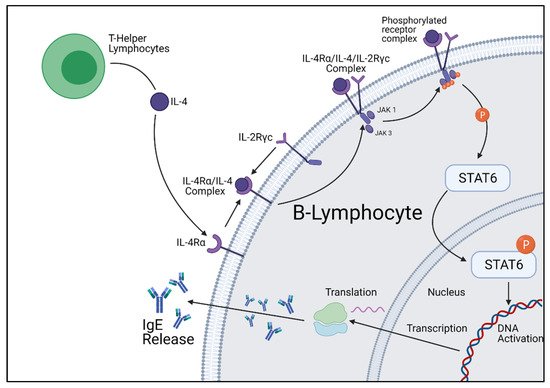
Figure 2. Cellular summary of B-lymphocyte IgE production initiated by IL-4 stimulation. Figure was created by the authors and reproduced with permission from BioRender.com (accessed on 19 November 2021).
3. IL-4 Pathway Inhibition
Due to their integral role in the production of IgE, the IL-4 and IL-13 cytokine pathways have been investigated as a target for potential therapeutic intervention. There exist a number of compounds that inhibit aspects of these cytokine pathways, although the only one that has been developed into a therapeutic that is approved for clinical use is dupilumab, a monoclonal antibody therapy that targets the IL-4 and IL-13 membrane receptors, specifically the alpha subunit of the IL-4Rα receptor. The IL-4Rα receptor is involved in both cytokine pathways, thereby making it the prime target for therapeutic inhibition. Dupilumab is currently approved for clinical use in the treatment of severe atopic dermatitis, though this treatment is invasive, requiring regular subdermal injections. There have been investigations into the use of dupilumab for the treatment of asthma, displaying promising results [17]. In one such study, conducted by Castro et al. in 2018, severe asthmatic patients were treated with either dupilumab or a placebo. Their results displayed that dupilumab had a significant positive effect in the treatment of asthma, reducing hospitalization visits and asthmatic episodes [18]. With no adverse side effects observed, dupilumab has a promising future, with ongoing studies evaluating and confirming its safety [19]. While only preliminary studies, these trials further solidify that IL-4- and IL-13-related inflammation are directly linked to asthma, and can be a target for therapeutic intervention [20].
Another drug that targets the IL-4 pathway is pascolizumab. This therapeutic is a humanized monoclonal antibody that targets the IL-4 cytokine. Initial animal studies were very promising, displaying almost complete T-cell function inhibition. Studies around this drug showed that pascolizumab had a high binding affinity to IL-4 with a slow dissociation rate [21]. This was coupled with low toxicity and few side effects, although these effects did become more exaggerated after chronic use, due to the drug’s long half-life. Previous animal trials have been conducted to evaluate short- and long-term treatment safety. The results from these trials were very promising, with the only complication being the development of an anti-idiotypic response resulting in the rapid clearance of pascolizumab [21]. In more recent years, a phase II clinical trial with pascolizumab was conducted with asthmatic patients. While no issues were raised with the safety of the treatment, the trials were discontinued due to the low efficacy [22].
Unlike the other therapeutics discussed, pitrakinra is a molecular therapeutic currently being investigated for IL-4 pathway inhibition. This peptide is a molecular inhibitor of the IL-4Rα surface receptor, competing for binding against both IL-4 and IL-13 [23]. Previous trials have shown pitrakinra having a low effect; however, as the trials progressed, pitrakinra began showing a greatly increased efficacy [24]. After further investigation, this variation was explained through genetic sequence variations between the patient groups, specifically a single nucleotide polymorphism within the IL-4Rα gene [25]. Results showed that patients carrying this SNP had a higher success rate than those without. Currently, pitrakinra is being further investigated for therapeutic effects within trials. Pitrakinra is a prime example of the role that genetics play in pharmaceuticals, displaying how a single nucleotide polymorphism can greatly affect the efficacy of a therapeutic. Further phenotypic investigations of the IL-4 and IL-13 pathways are required; this could allow for the subgrouping of patients by their genetic variations, opening the way to personalized therapeutic inhibitions.
References
- Mims, J.W. Asthma: Definitions and pathophysiology. Int. Forum Allergy Rhinol. 2015, 5 (Suppl. 1), S2–S6.
- Bagnasco, D.; Ferrando, M.; Varricchi, G.; Passalacqua, G.; Canonica, G.W. A Critical Evaluation of Anti-IL-13 and Anti-IL-4 Strategies in Severe Asthma. Int. Arch. Allergy Immunol. 2016, 170, 122–131.
- Sullivan, P.W.; Campbell, J.D.; Ghushchyan, V.H.; Globe, G. Outcomes before and after treatment escalation to Global Initiative for Asthma steps 4 and 5 in severe asthma. Ann. Allergy Asthma Immunol. 2015, 114, 462–469.
- Bateman, E.D.; Boushey, H.A.; Bousquet, J.; Busse, W.W.; Clark, T.J.; Pauwels, R.A.; Pedersen, S.E.; GOAL Investigators Group. Can guideline-defined asthma control be achieved? The Gaining Optimal Asthma ControL study. Am. J. Respir. Crit. Care Med. 2004, 170, 836–844.
- Gieseck, R.L.; Wilson, M.S.; Wynn, T.A. Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol. 2018, 18, 62–76.
- Bao, K.; Reinhardt, R.L. The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. Cytokine 2015, 75, 25–37.
- Zhu, J. T helper 2 (Th2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production. Cytokine 2015, 75, 14–24.
- Antczak, A.; Domanska-Senderowska, D.; Gorski, P.; Pastuszak-Lewandoska, D.; Nielepkowicz-Gozdzinska, A.; Szewczyk, K.; Kurmanowska, Z.; Kiszałkiewicz, J.; Brzeziańska-Lasota, E. Analysis of changes in expression of IL-4/IL-13/STAT6 pathway and correlation with the selected clinical parameters in patients with atopic asthma. Int. J. Immunopathol. Pharmacol. 2016, 29, 195–204.
- Kelly-Welch, A.; Hanson, E.M.; Keegan, A.D. Interleukin-4 (IL-4) pathway. Sci. Signal. 2005, 2005, cm9.
- Sheikh, F.; Dickensheets, H.; Pedras-Vasconcelos, J.; Ramalingam, T.; Helming, L.; Gordon, S.; Donnelly, R.P. The Interleukin-13 Receptor-α1 Chain Is Essential for Induction of the Alternative Macrophage Activation Pathway by IL-13 but Not IL-4. J. Innate Immun. 2015, 7, 494–505.
- Kelly-Welch, A.; Hanson, E.M.; Keegan, A.D. Interleukin-13 (IL-13) pathway. Sci. Signal. 2005, 2005, cm8.
- Wu, W.J.; Wang, S.H.; Wu, C.C.; Su, Y.A.; Chiang, C.Y.; Lai, C.H.; Wang, T.H.; Cheng, T.-L.; Kuo, J.-Y.; Hsu, T.-C.; et al. IL-4 and IL-13 Promote Proliferation of Mammary Epithelial Cells through STAT6 and IRS-1. Int. J. Mol. Sci. 2021, 22, 12008.
- Liu, C.; Richard, K.; Wiggins, M.; Zhu, X.; Conrad, D.H.; Song, W. CD23 can negatively regulate B-cell receptor signaling. Sci. Rep. 2016, 6, 25629.
- Kneitz, C.; Goller, M.; Seggewiss, R.; Yaman, A.; Serfling, E.; Tony, H.P. STAT6 and the regulation of CD23 expression in B-chronic lymphocytic leukemia. Leuk. Res. 2000, 24, 331–337.
- Walford, H.H.; Doherty, T.A. STAT6 and lung inflammation. JAK-STAT 2013, 2, e25301.
- Singh, D.; Ravi, A.; Southworth, T. CRTH2 antagonists in asthma: Current perspectives. Clin. Pharmacol. 2017, 9, 165–173.
- Wenzel, S.; Castro, M.; Corren, J.; Maspero, J.; Wang, L.; Zhang, B.; Pirozzi, G.; Sutherland, E.R.; Evans, R.R.; Joish, V.N.; et al. Dupilumab efficacy and safety in adults with uncontrolled persistent asthma despite use of medium-to-high-dose inhaled corticosteroids plus a long-acting β2 agonist: A randomised double-blind placebo-controlled pivotal phase 2b dose-ranging trial. Lancet 2016, 388, 31–44.
- Castro, M.; Corren, J.; Pavord, I.D.; Maspero, J.; Wenzel, S.; Rabe, K.F.; Busse, W.W.; Ford, L.; Sher, L.; FitzGerald, J.M.; et al. Dupilumab Efficacy and Safety in Moderate-to-Severe Uncontrolled Asthma. N. Engl. J. Med. 2018, 378, 2486–2496.
- McGregor, M.C.; Krings, J.G.; Nair, P.; Castro, M. Role of Biologics in Asthma. Am. J. Respir. Crit. Care Med. 2019, 199, 433–445.
- Santini, G.; Mores, N.; Malerba, M.; Mondino, C.; Anzivino, R.; Macis, G.; Montuschi, P. Dupilumab for the treatment of asthma. Expert Opin. Investig. Drugs 2017, 26, 357–366.
- Hart, T.K.; Blackburn, M.N.; Brigham-Burke, M.; Dede, K.; Al-Mahdi, N.; Zia-Amirhosseini, P.; Cook, R.M. Preclinical efficacy and safety of pascolizumab (SB 240683): A humanized anti-interleukin-4 antibody with therapeutic potential in asthma. Clin. Exp. Immunol. 2002, 130, 93–100.
- Long, A.A. Monoclonal antibodies and other biologic agents in the treatment of asthma. MAbs 2009, 1, 237–246.
- Antoniu, S.A. Pitrakinra, a dual IL-4/IL-13 antagonist for the potential treatment of asthma and eczema. Curr. Opin. Investig. Drugs 2010, 11, 1286–1294.
- Slager, R.E.; Otulana, B.A.; Hawkins, G.A.; Yen, Y.P.; Peters, S.P.; Wenzel, S.E.; Meyers, D.A.; Bleecker, R. IL-4 receptor polymorphisms predict reduction in asthma exacerbations during response to an anti-IL-4 receptor alpha antagonist. J. Allergy Clin. Immunol. 2012, 130, 516–522.e4.
- Pennington, E.J.; Wechsler, M.E.; Ortega, V.E. Chapter 9—Pharmacogenomics and Applications to Asthma Management. In Personalizing Asthma Management for the Clinician; Szefler, S.J., Holguin, F., Wechsler, M.E., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 97–112.
More
Information
Subjects:
Allergy
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
974
Revisions:
2 times
(View History)
Update Date:
11 Jan 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




