Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Wiwin Effendi | + 2113 word(s) | 2113 | 2021-12-28 07:36:59 | | | |
| 2 | Camila Xu | Meta information modification | 2113 | 2022-01-10 03:47:06 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Effendi, W. Hedgehog Signaling Pathway. Encyclopedia. Available online: https://encyclopedia.pub/entry/17916 (accessed on 04 March 2026).
Effendi W. Hedgehog Signaling Pathway. Encyclopedia. Available at: https://encyclopedia.pub/entry/17916. Accessed March 04, 2026.
Effendi, Wiwin. "Hedgehog Signaling Pathway" Encyclopedia, https://encyclopedia.pub/entry/17916 (accessed March 04, 2026).
Effendi, W. (2022, January 08). Hedgehog Signaling Pathway. In Encyclopedia. https://encyclopedia.pub/entry/17916
Effendi, Wiwin. "Hedgehog Signaling Pathway." Encyclopedia. Web. 08 January, 2022.
Copy Citation
The hedgehog (Hh) pathway is a sophisticated conserved cell signaling pathway that plays an essential role in controlling cell specification and proliferation, survival factors, and tissue patterning formation during embryonic development.
cell signaling
signal transduction
hedgehog pathway
1. Introduction
Cell signaling is a multifactorial system that represents the knot-like schematics of the signaling cascades that are used to transfer messages from the first messenger to the receptor and decoded through the signaling intermediates of the second messengers [1][2]. Signal transduction, as an aspect of cell signaling, describes how cells interpret and react to external events [3]. Hh is one of the major signal transduction networks for intercellular communication during embryonic development and organogenesis [4] and it regulates mitogenic and morphogenic functions during organ development [5]. Nevertheless, many disease processes arise from defects or through the aberrant activation of these developmental pathways.
The dysregulation of the Hh signaling network in controlled cell growth and division induces autocrine and paracrine function distortions, leading to the development of tumorigenesis and cancer progression [6][7]. Myofibroblast-associated Hh signaling is involved in accelerated tumor growth in various cancers [8][9][10]. Moreover, Hh signaling is also responsible for the development of numerous lung diseases [11]. Recent gene expression studies and animal disease models have demonstrated that Hh signaling can induce the fibroblast to myofibroblast transition (myofibroblast differentiation) in IPF [12].
2. Hh Signal Transduction
Vertebrate genome duplication categorized Hh genes into three different types of hedgehog proteins: the Desert Hedgehog (Dhh), Indian Hedgehog (Ihh), and Sonic Hedgehog (Shh) proteins [13].
2.1. Element of Hh Signal Transduction
Hh proteins undergo multiple processing steps that are required for the generation and release of the active ligand from the producing cell. The core components that mediate the Hh signal response in vertebrae are two patched receptors (Ptch1, Ptch2), a key signal transducer smoothened receptor (Smo), three glioma-associated oncogene (Gli) transcription factors (Gli1, Gli2, Gli3), the suppressed fusion homolog (Sufu), and kinesine protein 7 (Kif7) [14].
2.1.1. Hh Ligand
Hh genes are automatically cleaved into a 20 kDa N-terminal protein (Hh-N) and a 25 kDa C-terminal protein (Hh-C) [15]. After translation, the Hh protein is then transported to the endoplasmic reticulum for dual lipid modification. The first modification removes the C-terminal domain and attaches cholesterol to the C-terminal (the C-terminally cholesterol-modified N-terminal Hh signaling domain (HhN)), leading to the association of Hh with membranes [16]. Next, a palmitate molecule is attached to the N-terminal by Hh acyltransferase (Hhat), resulting in a fully active dual lipid-modified HhNp [17]. The dual lipid-modified HhNp is then transported to the cell surface.
2.1.2. Ptch
Hh ligands start to trigger signaling in the target cells by binding a 12-pass integral membrane, the Ptch protein (complex Hh-Ptch). Vertebrates have two Ptch genes, Ptch1 and Ptch2, but Ptch1 is the primary signaling regulator [18]. Ptch1 is essential for Hh signaling and for generating stable signaling gradients due to negative feedback, the inhibition of Hh ligands and Smo, and involvement in a double-negative circuit (in which Hh inhibits Ptch and also blocks Smo). [19]. The Ptch1 protein has a sterol-sensing domain (SSD), two large extracellular loops, and a C-terminal cytoplasmic tail [20]. SSD mediates the vesicular trafficking of Ptch1 to regulate Smo activity [21].
2.1.3. Smo
The G protein-coupled receptor (GPCR), the Smo protein, which is predominantly located in the membrane of intracellular endosomes, functions as a co-receptor and a positive regulator of the Hh signaling pathway. Smo consists of an amino-terminal cysteine-rich domain (CRD), three extracellular and three intracellular loops (ECL and ICL), seven transmembrane domains (TM), and an intracellular carboxyl-terminal tail that is able to undergo a range of post-translational modifications [22].
2.1.4. Gli
The Gli family of latent zinc-finger proteins function as transcriptional mediators and are implicated in the activation and repression of the Hh target genes [23]. In detail, Gli1 only acts as a transcriptional activator (GliA), Gli2 is the principal Hh-regulated transcriptional activator, and Gli3 is the strongest Hh-regulated repressor (GliR) [24]. The differential activity of Gli is regulated on the level of ubiquitin-mediated proteolytic processing [25] and the subcellular localization of a nuclear localization signal (NLS) and a nuclear export signal (NES) [26].
2.1.5. Sufu
Sufu is an essential intracellular negative regulator of Hh signaling and acts by binding and modulating the Gli transcription factors [27]. In the absence of signaling, Sufu inhibits the Gli transcription factors by binding Gli through a C-terminal Sufu-interacting site (SIC) that is responsible for the Sufu-mediated cytoplasmic retention of Gli1 via the N-terminal Sufu-interacting (SIN) pathway [28].
2.1.6. Kif7
2.2. Hh Signaling Pathway
The Hh activates canonical (either through ligand-dependent interaction or receptor-induced signaling) and non-canonical (ligand-independent interaction) signaling pathways (Figure 1).
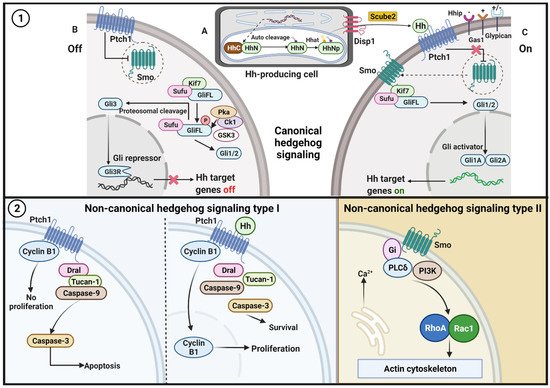
Figure 1. Hh pathway. (1) Canonical signaling. A. The production and secretion of Hh ligands/proteins. After translation, the precursor of the Hh protein is transported to the endoplasmic reticulum (ER) for autoclaved and dual-lipid modification. The first modification replaces the C-terminal domain from Hh-N with cholesterol at the C terminus; then, a palmitate molecule is attached to the N-terminal Hh-N by the Hh acyltransferase (Hhat). Disp1 on the cell surface and Scube2 regulates Hh-N secretion and distribution into the extracellular space of the producing cells. B. In the absence of the Hh ligands, Ptch functions to suppress any inactive Smo that is inside the cell and inhibits the migration of Smo to the membrane. Sufu restrains the GliFL protein in the cytoplasm, and GliFL is then phosphorylated at multiple sites in the C-terminal region by PKA, GSK3, and CK1. Next, the truncated GliR (Gli3) translocates to the nucleus and binds to the Hh target gene (target gene off). C. Ptch1 binds to the Hh ligand and releases the Smo that has been inhibited by Ptch. Active Smo induces the release of GliFL from cytoplasmic retention. The Hh–Ptch1 complex is then internalized and is degraded in the lysosomes. In the end, GliFL is converted to its active form GliA (Gli1 and Gli2) and migrates to the nucleus to activate several target genes (target gene on). Coreceptors for the Hh ligand pathway activate positive regulators (Gas1), negative regulators (Hhip), and dual functions (Glypican). (2) Non-canonical signaling. Type I, in the absence of Hh ligand, Ptch recruits complex proteins Dral, Tucan-1, and caspase-9 that followed by caspase-3 activation, which further amplifies cell apoptosis. The binding of Hh ligand to Ptch disorganizes the interaction of Cyclin B1 and the proapoptotic complex, leading to increased proliferation and survival. Type II, activation of Smo leads to dissociation of Gi, activation of PI3K and RhoA and Rac1, which then modulate the actin cytoskeleton and induce elevation of intracellular calcium.
2.2.1. Canonical Pathway
The canonical signaling pathways focus on the mechanism by which Hh regulates the Gli [32]. This pathway can be operated in both the presence and absence of Hh ligands.
In the absence of Hh ligands, Ptch1 functions to suppress any inactive Smo that is inside the cell and inhibits the migration of Smo to the membrane [33]. The mechanism through which Ptch1 inhibits Smo is not precise. It is supposed that Ptch1 is transported out of the cell as an endogenous intracellular small molecule that acts as an agonist for Smo and that does not bind to Smo [34]. Ptch1 requires extracellular Na+ and membrane cholesterol to regulate Smo [35]. Furthermore, Ptch1 might inhibit Smo through an indirect mechanism, possibly through changes in the distribution or concentration of a small molecule [36].
The inhibition of Smo activity is an essential step for activating this pathway in mammals [37]. The full-length Gli (GliFL) is then phosphorylated at multiple sites in the C-terminal region by protein kinase A (PKA), glycogen synthase kinase-3 (GSK3), and casein kinase 1 (CK1) [38]. Kif7 acts as a scaffolding protein for PKA, GSK3, and CK1 during the Gli phosphorylation [32]. Without Hh, Sufu restrains the GliFL protein in the cytoplasm, whereas ligand binding will proteolytically cleave Gli from Sufu [39]. The truncated GliR (Gli3) then translocates to the nucleus and binds to the Hh target gene [40].
The Ptch1 protein stops inhibiting Smo after binding the Hh ligand and limits the half-life of the ligand [41]. Smo activation induces the stabilization and release of Gli, the transducer of the significant cellular effects of canonical Hh signaling, from cytoplasmic retention [42]. The Hh–Ptch1 complex is then internalized and degraded in the lysosomes [43]. Hh signaling is subsequently activated and transmitted via a protein complex that includes Kif7 and Sufu [44]. Finally, GliFL is converted to its active form GliA (Gli1 and Gli2) and migrates to the nucleus to activate several target genes [45]. Canonical Hh signaling leads to Gli code regulation, which covers the sum of all of the positive and negative functions of all of the Gli proteins [40].
2.2.2. Non-Canonical Pathway
Contrary to canonical complex signaling network resulting in activation of the Gli family of transcription factors, some Hh signaling proceeds through Gli independent activation. In detail, non-canonical Hh delivers signals via (1) Ptch1 in the presence of Hh ligand, (2) Ptch1 in the absence of Hh ligand, and (3) Smo-dependent and Gi protein modulating Ca2+ and actin skeleton [32][46][47][48].
After embryogenesis, the Hh pathway continues to signal to discrete populations of stem and progenitor cells within various organs in order to maintain tissue homeostasis and repair [49]. In the lungs, the Hh pathway never entirely disappears from development to adulthood, but the activation domain shifts dramatically and repurposes itself in order to maintain cellular homeostasis and organ function [50]. It seems that the Hh pathway is silenced until it is reactivated by tissue injury in order to mediate cellular regeneration and repair.
3. Hh Signaling in Lung Development
It is already known that the Hh signaling pathway plays a critical role as the principal regulator in the normal development of many tissues such as those in the lung. Lung morphogenesis relies on intricate interactions and the coordinated development of the endoderm layer-derived epithelial cells into the surrounding mesoderm-derived mesenchyme [51].
Embryonic lung development follows the principle of branching morphogenesis into five phases; the first four phases (embryonic, pseudo glandular, canalicular, and saccular) result in a typical branching structure that ends with alveolar sacs with a surrounding stromal scaffold and vascular structures where during the final (postnatal) alveolar phase, the terminal sacs give rise to mature alveolar ducts and alveoli [11]. The first two development stages regulate the establishment of the conducting airways, and the last three stages are responsible for vascular development, alveolar development, and reducing mesenchymal tissue, which is crucial for the formation of the thin air–blood interface that is indispensable for gas exchange [52]. Hh signaling is a crucial aspect that can be used to orchestrate a network of growth factors, transcription factors, and extracellular matrix molecules during lung embryogenesis [11].
During embryogenesis, the Shh that is secreted by the epithelial cells during the early steps of embryogenesis, act as a spatial regulator of bronchial bud formation and are essential for the mesenchymal–epithelial cross-talk that guides branching and epithelial tube elongation, as well as smooth muscle cell/myofibroblast differentiation [53]. Other elements of Hh signaling, such as Ptch1, Smo, and Gli1-3, are mainly expressed in the epithelium but are expressed weakly in the mesenchyme of the developing human lung [54]. Inhibition of the Shh pathway in mouse models causes severe lung malformations, resulting in hypoplasia and tracheal malformations and non-viable phenotypes [55]. He et al. demonstrated that Shh signaling controlled multiple morphogen signaling pathways, such as Fgf10 expression, in lung morphogenesis via heparan sulfate (HS) glycosaminoglycans [56].
In contrast with its crucial roles during embryonic development, Hh signaling has more restricted roles after birth. Postnatally, mature lung development begins with the formation of the alveolar septum (alveolarization) followed by secondary septa and microvascular maturation [57]. The Hh pathway also regulates mesenchymal proliferation and myofibroblast function during the septum alveolarization and maturation phase [58]. Overall, Hh signaling plays a vital role in lung embryogenesis, homeostasis, and regeneration via the fine cellular distribution of the Hh pathway components, which orchestrate complex cross-talk between lung cell populations, leading to proper lung development [59].
Recent studies indicate that the growth signaling pathways may be reactivated in tissue remodeling and cancer development. IPF and lung cancer share similar cellular and molecular pathological processes, including aberrant embryological pathways [60]. Hh signaling is one of the pathways that is responsible for the activation and proliferation of both the myofibroblasts in IPF and cancer-associated fibroblasts (CAF) [61].
Several studies showed the involvement of the Hh pathway during fibroblast activation and during myofibroblast transformation in biliary and liver fibrosis [62][63][64][65] as well as in kidney fibrosis [66][67]. Froidure et al. proposed that minimal aberrance in Hh signaling could induce the development and progression of pulmonary fibrosis rather than repair in a chronically injured lung [68]. However, a recent study declared that overexpression of Hh signaling in diabetic myocardial ischemia reduces cardiac fibrosis via suppressed myocardial apoptosis and improved myocardial angiogenesis [69].
References
- Nair, A.; Chauhan, P.; Saha, B.; Kubatzky, K.F. Conceptual Evolution of Cell Signaling. Int. J. Mol. Sci. 2019, 20, 3292.
- Di-Bella, J.P.; Colman-Lerner, A.; Ventura, A.C. Properties of cell signaling pathways and gene expression systems operating far from steady-state. Sci. Rep. 2018, 8, 1–14.
- Handly, L.N.; Yao, J.; Wollman, R. Signal Transduction at the Single-Cell Level: Approaches to Study the Dynamic Nature of Signaling Networks. J. Mol. Biol. 2016, 428, 3669–3682.
- Ryan, K.E.; Chiang, C. Hedgehog secretion and signal transduction in vertebrates. J. Biol. Chem. 2012, 287, 17905–17913.
- Groves, I.; Placzek, M.; Fletcher, A.G. Of mitogens and morphogens: Modelling Sonic Hedgehog mechanisms in vertebrate development. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190660.
- Sever, R.; Brugge, J.S. Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 2015, 5, a006098.
- Niyaz, M.; Khan, M.S.; Mudassar, S. Hedgehog Signaling: An Achilles’ Heel in Cancer. Transl. Oncol. 2019, 12, 1334–1344.
- Domenech, M.; Bjerregaard, R.; Bushman, W.; Beebe, D.J. Hedgehog signaling in myofibroblasts directly promotes prostate tumor cell growth. Integr. Biol. 2012, 4, 142–152.
- Zhang, J.; Fan, J.; Zeng, X.; Nie, M.; Luan, J.; Wang, Y.; Ju, D.; Yin, K. Hedgehog signaling in gastrointestinal carcinogenesis and the gastrointestinal tumor microenvironment. Acta Pharm. Sin. B 2021, 11, 609–620.
- Steele, N.G.; Biffi, G.; Kemp, S.B.; Zhang, Y.; Drouillard, D.; Syu, L.; Hao, Y.; Oni, T.E.; Brosnan, E.; Elyada, E.; et al. Inhibition of Hedgehog Signaling Alters Fibroblast Composition in Pancreatic Cancer. Clin. Cancer Res. 2021, 27, 2023–2037.
- Kugler, M.C.; Joyner, A.L.; Loomis, C.A.; Munger, J.S. Sonic hedgehog signaling in the lung. From development to disease. Am. J. Respir. Cell Mol. Biol. 2015, 52, 1–13.
- Chen, X.; Shi, C.; Cao, H.; Chen, L.; Hou, J.; Xiang, Z.; Hu, K.; Han, X. The hedgehog and Wnt/β-catenin system machinery mediate myofibroblast differentiation of LR-MSCs in pulmonary fibrogenesis. Cell Death Dis. 2018, 9, 639.
- Varjosalo, M.; Taipale, J. Hedgehog: Functions and mechanisms. Genes Dev. 2008, 22, 2454–2472.
- Jia, Y.; Wang, Y.; Xie, J. The Hedgehog pathway: Role in cell differentiation, polarity and proliferation. Arch. Toxicol. 2015, 89, 179–191.
- Bürglin, T.R. The Hedgehog protein family. Genome Biol. 2008, 9, 241.
- Varjosalo, M.; Taipale, J. Hedgehog signaling. J. Cell Sci. 2007, 120, 3–6.
- Buglino, J.A.; Resh, M.D. Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J. Biol. Chem. 2008, 283, 22076–22088.
- Nieuwenhuis, E.; Hui, C.C. Hedgehog signaling and congenital malformations. Clin. Genet. 2005, 67, 193–208.
- Kong, J.H.; Siebold, C.; Rohatgi, R. Biochemical mechanisms of vertebrate hedgehog signaling. Development 2019, 146, dev166892.
- Kawamura, S.; Hervold, K.; Ramirez-Weber, F.A.; Kornberg, T.B. Two patched protein subtypes and a conserved domain of group I proteins that regulates turnover. J. Biol. Chem. 2008, 283, 30964–30969.
- Martín, V.; Carrillo, G.; Torroja, C.; Guerrero, I. The sterol-sensing domain of patched protein seems to control smoothened activity through patched vesicular trafficking. Curr. Biol. 2001, 11, 601–607.
- Arensdorf, A.M.; Marada, S.; Ogden, S.K. Smoothened Regulation: A Tale of Two Signals. Trends Pharmacol. Sci. 2016, 37, 62–72.
- Koebernick, K.; Pieler, T. Gli-type zinc finger proteins as bipotential transducers of Hedgehog signaling. Differentiation 2002, 70, 69–76.
- Niewiadomski, P.; Niedziółka, S.M.; Markiewicz, Ł.; Uśpieński, T.; Baran, B.; Chojnowska, K. Gli Proteins: Regulation in Development and Cancer. Cells 2019, 8, 147.
- Jiang, J. Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell Cycle 2006, 5, 2457–2463.
- Hatayama, M.; Aruga, J. Gli Protein Nuclear Localization Signal. In Vitamins and Hormones; Academic Press Inc.: Cambridge, MA, USA, 2012; Volume 88, pp. 73–89.
- Cherry, A.L.; Finta, C.; Karlström, M.; Jin, Q.; Schwend, T.; Astorga-Wells, J.; Zubarev, R.A.; Del Campo, M.; Criswell, A.R.; De Sanctis, D.; et al. Structural basis of SUFU-GLI interaction in human Hedgehog signalling regulation. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 2563–2579.
- Han, Y.; Shi, Q.; Jiang, J. Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc. Natl. Acad. Sci. USA 2015, 112, 6383–6388.
- Liem, K.F.; He, M.; Ocbina, P.J.R.; Anderson, K.V. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 13377–13382.
- Hsu, S.H.C.; Zhang, X.; Yu, C.; Li, Z.J.; Wunder, J.S.; Hui, C.C.; Alman, B.A. Kif7 promotes hedgehog signaling in growth plate chondrocytes by restricting the inhibitory function of Sufu. Development 2011, 138, 3791–3801.
- Maurya, A.K.; Ben, J.; Zhao, Z.; Lee, R.T.H.; Niah, W.; Ng, A.S.M.; Iyu, A.; Yu, W.; Elworthy, S.; van Eeden, F.J.M.; et al. Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo. PLoS Genet. 2013, 9, e1003955.
- Robbins, D.J.; Fei, D.L.; Riobo, N.A. The hedgehog signal transduction network. Sci. Signal. 2012, 5, re6.
- Bijlsma, M.F.; Spek, C.A.; Zivkovic, D.; van de Water, S.; Rezaee, F.; Peppelenbosch, M.P. Repression of Smoothened by Patched-Dependent (Pro-)Vitamin D3 Secretion. PLoS Biol. 2006, 4, e232.
- Rubin, L.L.; de Sauvage, F.J. Targeting the Hedgehog pathway in cancer. Nat. Rev. Drug Discov. 2006, 5, 1026–1033.
- Myers, B.R.; Neahring, L.; Zhang, Y.; Roberts, K.J.; Beachy, P.A. Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc. Natl. Acad. Sci. USA 2017, 114, E11141–E11150.
- Taipale, J.; Cooper, M.K.; Maiti, T.; Beachy, P.A. Patched acts catalytically to suppress the activity of smoothened. Nature 2002, 418, 892–897.
- Sari, I.N.; Phi, L.T.H.; Jun, N.; Wijaya, Y.T.; Lee, S.; Kwon, H.Y. Hedgehog Signaling in Cancer: A Prospective Therapeutic Target for Eradicating Cancer Stem Cells. Cells 2018, 7, 208.
- Hui, C.C.; Angers, S. Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 2011, 27, 513–537.
- Humke, E.W.; Dorn, K.V.; Milenkovic, L.; Scott, M.P.; Rohatgi, R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010, 24, 670–682.
- Aberger, F.; Ruiz i Altaba, A. Context-dependent signal integration by the GLI code: The oncogenic load, pathways, modifiers and implications for cancer therapy. Semin. Cell Dev. Biol. 2014, 33, 93–104.
- Pak, E.; Segal, R.A. Hedgehog Signal Transduction: Key Players, Oncogenic Drivers, and Cancer Therapy. Dev. Cell 2016, 38, 333–344.
- Szczepny, A.; Rogers, S.; Jayasekara, W.S.N.; Park, K.; McCloy, R.A.; Cochrane, C.R.; Ganju, V.; Cooper, W.A.; Sage, J.; Peacock, C.D.; et al. The role of canonical and non-canonical Hedgehog signaling in tumor progression in a mouse model of small cell lung cancer. Oncogene 2017, 36, 5544–5550.
- Incardona, J.P.; Gruenberg, J.; Roelink, H. Sonic hedgehog induces the segregation of patched and smoothened in endosomes. Curr. Biol. 2002, 12, 983–995.
- He, M.; Subramanian, R.; Bangs, F.; Omelchenko, T.; Liem, K.F.; Kapoor, T.M.; Anderson, K.V. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol. 2014, 16, 663–672.
- Cohen, M.; Kicheva, A.; Ribeiro, A.; Blassberg, R.; Page, K.M.; Barnes, C.P.; Briscoe, J. Ptch1 and Gli regulate Shh signalling dynamics via multiple mechanisms. Nat. Commun. 2015, 6, 1–12.
- Carballo, G.B.; Honorato, J.R.; De Lopes, G.P.F.; Spohr, T.C.L.D.S.E. A highlight on Sonic hedgehog pathway. Cell Commun. Signal. 2018, 16, 1–15.
- Pietrobono, S.; Gagliardi, S.; Stecca, B. Non-canonical hedgehog signaling pathway in cancer: Activation of GLI transcription factors beyond smoothened. Front. Genet. 2019, 10, 1–20.
- Jeng, K.S.; Chang, C.F.; Lin, S.S. Sonic hedgehog signaling in organogenesis, tumors, and tumor microenvironments. Int. J. Mol. Sci. 2020, 21, 758.
- Petrova, R.; Joyner, A.L. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 2014, 141, 3445–3457.
- Wang, C.; Cassandras, M.; Peng, T. The role of Hedgehog signaling in adult lung regeneration and maintenance. J. Dev. Biol. 2019, 7, 14.
- McCulley, D.; Wienhold, M.; Sun, X. The pulmonary mesenchyme directs lung development. Curr. Opin. Genet. Dev. 2015, 32, 98–105.
- Fernandes-Silva, H.; Correia-Pinto, J.; Moura, R.S. Canonical Sonic Hedgehog Signaling in Early Lung Development. J. Dev. Biol. 2017, 5, 3.
- Yin, M.; Ahlbrecht, K.; Seeger, W.; Voswinckel, R. The role of Sonic Hedgehog in postnatal mouse lung development. Pneumologie 2012, 66, A404.
- Zhang, M.; Wang, H.; Teng, H.; Shi, J.; Zhang, Y. Expression of SHH signaling pathway components in the developing human lung. Histochem. Cell Biol. 2010, 134, 327–335.
- Giroux-Leprieur, E.; Costantini, A.; Ding, V.W.; He, B. Hedgehog Signaling in Lung Cancer: From Oncogenesis to Cancer Treatment Resistance. Int. J. Mol. Sci. 2018, 19, 2835.
- He, H.; Huang, M.; Sun, S.; Wu, Y.; Lin, X. Epithelial heparan sulfate regulates Sonic Hedgehog signaling in lung development. PLoS Genet. 2017, 13, 1–22.
- Burri, P.H. Structural aspects of postnatal lung development-alveolar formation and growth. Biol. Neonate 2006, 89, 313–322.
- Kugler, M.C.; Loomis, C.A.; Zhao, Z.; Cushman, J.C.; Liu, L.; Munger, J.S. Sonic Hedgehog Signaling Regulates Myofibroblast Function during Alveolar Septum Formation in Murine Postnatal Lung. Am. J. Respir. Cell Mol. Biol. 2017, 57, 280–293.
- Belgacemi, R.; Danopoulos, S.; Deslée, G.; Dormoy, V.; Al Alam, D. Hedgehog signalling crosstalks orchestrate human lung development. Eur. Respir. J. 2020, 56, 596.
- Ballester, B.; Milara, J.; Cortijo, J. Idiopathic Pulmonary Fibrosis and Lung Cancer: Mechanisms and Molecular Targets. Int. J. Mol. Sci. 2019, 20, 593.
- Samarelli, A.V.; Masciale, V.; Aramini, B.; Coló, G.P.; Tonelli, R.; Marchioni, A.; Bruzzi, G.; Gozzi, F.; Andrisani, D.; Castaniere, I.; et al. Molecular Mechanisms and Cellular Contribution from Lung Fibrosis to Lung Cancer Development. Int. J. Mol. Sci. 2021, 22, 12179.
- Omenetti, A.; Porrello, A.; Jung, Y.; Yang, L.; Popov, Y.; Choi, S.S.; Witek, R.P.; Alpini, G.; Venter, J.; Vandongen, H.M.; et al. Hedgehog signaling regulates epithelial-mesenchymal transition during biliary fibrosis in rodents and humans. J. Clin. Investig. 2008, 118, 3331–3342.
- Yang, J.-J.; Tao, H.; Li, J. Hedgehog signaling pathway as key player in liver fibrosis: New insights and perspectives. Expert Opin. Ther. Targets 2014, 18, 1011–1021.
- Shen, X.; Peng, Y.; Li, H. The Injury-Related Activation of Hedgehog Signaling Pathway Modulates the Repair-Associated Inflammation in Liver Fibrosis. Front. Immunol. 2017, 8, 1450.
- Gao, L.; Zhang, Z.; Zhang, P.; Yu, M.; Yang, T. Role of canonical Hedgehog signaling pathway in liver. Int. J. Biol. Sci. 2018, 14, 1636–1644.
- Edeling, M.; Ragi, G.; Huang, S.; Pavenstädt, H.; Susztak, K. Developmental signalling pathways in renal fibrosis: The roles of Notch, Wnt and Hedgehog. Nat. Rev. Nephrol. 2016, 12, 426–439.
- Zhou, D.; Tan, R.J.; Liu, Y. Sonic hedgehog signaling in kidney fibrosis: A master communicator. Sci. China Life Sci. 2016, 59, 920–929.
- Froidure, A.; Marchal-Duval, E.; Homps-Legrand, M.; Ghanem, M.; Justet, A.; Crestani, B.; Mailleux, A. Chaotic activation of developmental signalling pathways drives idiopathic pulmonary fibrosis. Eur. Respir. Rev. 2020, 29, 190140.
- Xiao, Q.; Zhao, X.; Jiang, R.; Chen, X.; Zhu, X.; Chen, K.; Chen, S.; Zhang, X.; Qin, Y.; Liu, Y.; et al. Increased expression of Sonic hedgehog restores diabetic endothelial progenitor cells and improves cardiac repair after acute myocardial infarction in diabetic mice. Int. J. Mol. Med. 2019, 44, 1091–1105.
More
Information
Subjects:
Allergy
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.2K
Revisions:
2 times
(View History)
Update Date:
10 Jan 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





